Magnesium »
PDB 4yp9-4z4g »
4ypm »
Magnesium in PDB 4ypm: Crystal Structure of A Lona Protease Domain in Complex with Bortezomib
Enzymatic activity of Crystal Structure of A Lona Protease Domain in Complex with Bortezomib
All present enzymatic activity of Crystal Structure of A Lona Protease Domain in Complex with Bortezomib:
3.4.21.53;
3.4.21.53;
Protein crystallography data
The structure of Crystal Structure of A Lona Protease Domain in Complex with Bortezomib, PDB code: 4ypm
was solved by
C.-C.Lin,
C.-I.Chang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.85 / 1.85 |
| Space group | P 6 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 122.343, 122.343, 103.125, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 17.7 / 20.1 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of A Lona Protease Domain in Complex with Bortezomib
(pdb code 4ypm). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of A Lona Protease Domain in Complex with Bortezomib, PDB code: 4ypm:
In total only one binding site of Magnesium was determined in the Crystal Structure of A Lona Protease Domain in Complex with Bortezomib, PDB code: 4ypm:
Magnesium binding site 1 out of 1 in 4ypm
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of A Lona Protease Domain in Complex with Bortezomib
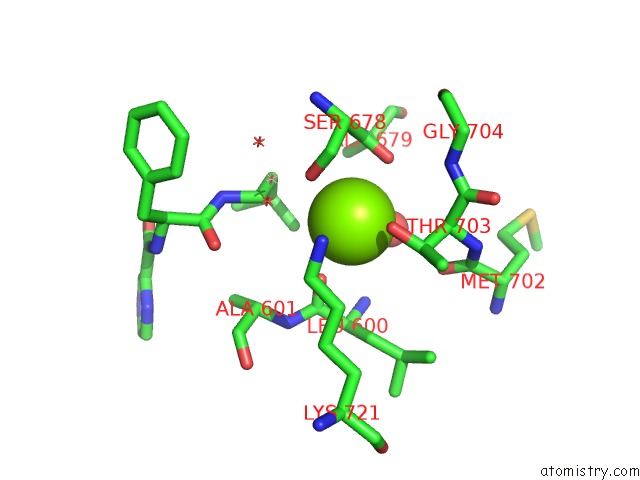
Mono view
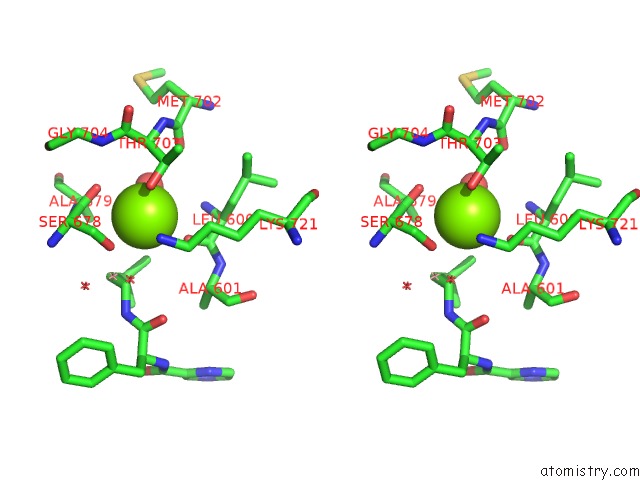
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of A Lona Protease Domain in Complex with Bortezomib within 5.0Å range:
|
Reference:
S.-C.Su,
C.-C.Lin,
H.-C.Tai,
M.-Y.Chang,
M.-R.Ho,
C.S.Babu,
J.-H.Liao,
S.-H.Wu,
Y.-C.Chang,
C.Lim,
C.-I.Chang.
Structural Basis For the Magnesium-Dependent Activation and Hexamerization of the Lon Aaa+ Protease Structure V. 24 676 2016.
ISSN: ISSN 0969-2126
PubMed: 27041593
DOI: 10.1016/J.STR.2016.03.003
Page generated: Tue Aug 12 04:23:25 2025
ISSN: ISSN 0969-2126
PubMed: 27041593
DOI: 10.1016/J.STR.2016.03.003
Last articles
Mg in 5JVHMg in 5K0N
Mg in 5K0L
Mg in 5JZQ
Mg in 5K0K
Mg in 5JYG
Mg in 5JZJ
Mg in 5JZD
Mg in 5JYS
Mg in 5JYD