Magnesium »
PDB 5btm-5c25 »
5bvc »
Magnesium in PDB 5bvc: Crystal Structure of Lipomyces Starkeyi Levoglucosan Kinase Bound to Adp, Magnesium and Levoglucosan in An Alternate Orientation.
Protein crystallography data
The structure of Crystal Structure of Lipomyces Starkeyi Levoglucosan Kinase Bound to Adp, Magnesium and Levoglucosan in An Alternate Orientation., PDB code: 5bvc
was solved by
J.P.Bacik,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.27 / 2.00 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 70.250, 70.250, 264.190, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.4 / 23.2 |
Magnesium Binding Sites:
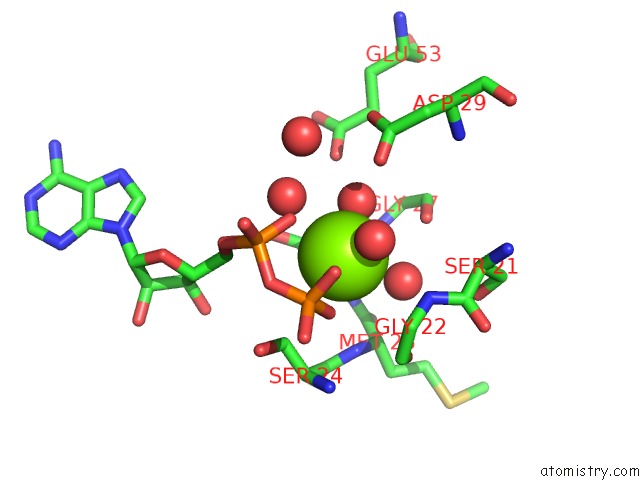
The binding sites of Magnesium atom in the Crystal Structure of Lipomyces Starkeyi Levoglucosan Kinase Bound to Adp, Magnesium and Levoglucosan in An Alternate Orientation.
(pdb code 5bvc). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of Lipomyces Starkeyi Levoglucosan Kinase Bound to Adp, Magnesium and Levoglucosan in An Alternate Orientation., PDB code: 5bvc:
In total only one binding site of Magnesium was determined in the Crystal Structure of Lipomyces Starkeyi Levoglucosan Kinase Bound to Adp, Magnesium and Levoglucosan in An Alternate Orientation., PDB code: 5bvc:
Magnesium binding site 1 out of 1 in 5bvc
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of Lipomyces Starkeyi Levoglucosan Kinase Bound to Adp, Magnesium and Levoglucosan in An Alternate Orientation.
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Lipomyces Starkeyi Levoglucosan Kinase Bound to Adp, Magnesium and Levoglucosan in An Alternate Orientation. within 5.0Å range:
|
Reference:
J.P.Bacik,
J.R.Klesmith,
T.A.Whitehead,
L.R.Jarboe,
C.J.Unkefer,
B.L.Mark,
R.Michalczyk.
Producing Glucose 6-Phosphate From Cellulosic Biomass: Structural Insights Into Levoglucosan Bioconversion. J.Biol.Chem. V. 290 26638 2015.
ISSN: ESSN 1083-351X
PubMed: 26354439
DOI: 10.1074/JBC.M115.674614
Page generated: Sun Sep 29 01:44:53 2024
ISSN: ESSN 1083-351X
PubMed: 26354439
DOI: 10.1074/JBC.M115.674614
Last articles
Fe in 9J2FFe in 9JDC
Fe in 9JDB
Fe in 9KU6
Fe in 9F47
Fe in 9F46
Fe in 9FSR
Fe in 9FSQ
Fe in 9FSS
Fe in 9FSP