Magnesium »
PDB 5btn-5c28 »
5bwm »
Magnesium in PDB 5bwm: The Complex Structure of C3CER Exoenzyme and Gdp Bound Rhoa (Nadh- Bound State)
Protein crystallography data
The structure of The Complex Structure of C3CER Exoenzyme and Gdp Bound Rhoa (Nadh- Bound State), PDB code: 5bwm
was solved by
A.Toda,
T.Tsurumura,
T.Yoshida,
H.Tsuge,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.56 / 2.50 |
| Space group | P 32 |
| Cell size a, b, c (Å), α, β, γ (°) | 50.479, 50.479, 136.671, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 20.9 / 24.6 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the The Complex Structure of C3CER Exoenzyme and Gdp Bound Rhoa (Nadh- Bound State)
(pdb code 5bwm). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the The Complex Structure of C3CER Exoenzyme and Gdp Bound Rhoa (Nadh- Bound State), PDB code: 5bwm:
In total only one binding site of Magnesium was determined in the The Complex Structure of C3CER Exoenzyme and Gdp Bound Rhoa (Nadh- Bound State), PDB code: 5bwm:
Magnesium binding site 1 out of 1 in 5bwm
Go back to
Magnesium binding site 1 out
of 1 in the The Complex Structure of C3CER Exoenzyme and Gdp Bound Rhoa (Nadh- Bound State)
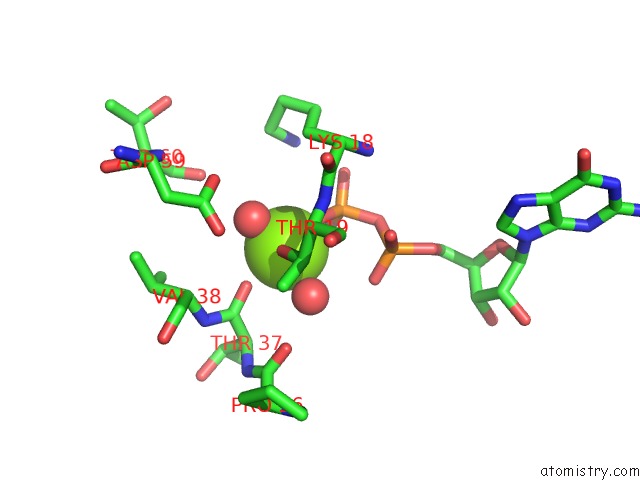
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of The Complex Structure of C3CER Exoenzyme and Gdp Bound Rhoa (Nadh- Bound State) within 5.0Å range:
|
Reference:
A.Toda,
T.Tsurumura,
T.Yoshida,
Y.Tsumori,
H.Tsuge.
Rho Gtpase Recognition By C3 Exoenzyme Based on C3-Rhoa Complex Structure. J.Biol.Chem. V. 290 19423 2015.
ISSN: ESSN 1083-351X
PubMed: 26067270
DOI: 10.1074/JBC.M115.653220
Page generated: Sun Sep 29 01:45:28 2024
ISSN: ESSN 1083-351X
PubMed: 26067270
DOI: 10.1074/JBC.M115.653220
Last articles
F in 7K1HF in 7K2U
F in 7K1F
F in 7K1E
F in 7K1I
F in 7K1D
F in 7JY4
F in 7K0V
F in 7JYM
F in 7K15