Magnesium »
PDB 5c29-5ca1 »
5c46 »
Magnesium in PDB 5c46: Crystal Structure of An Engineered Construct of Phosphatidylinositol 4 Kinase III Beta in Complex with Gtp Gamma S Loaded RAB11
Enzymatic activity of Crystal Structure of An Engineered Construct of Phosphatidylinositol 4 Kinase III Beta in Complex with Gtp Gamma S Loaded RAB11
All present enzymatic activity of Crystal Structure of An Engineered Construct of Phosphatidylinositol 4 Kinase III Beta in Complex with Gtp Gamma S Loaded RAB11:
2.7.1.67;
2.7.1.67;
Protein crystallography data
The structure of Crystal Structure of An Engineered Construct of Phosphatidylinositol 4 Kinase III Beta in Complex with Gtp Gamma S Loaded RAB11, PDB code: 5c46
was solved by
J.E.Burke,
M.L.Fowler,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.98 / 2.65 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 48.940, 97.950, 190.430, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.6 / 24.6 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of An Engineered Construct of Phosphatidylinositol 4 Kinase III Beta in Complex with Gtp Gamma S Loaded RAB11
(pdb code 5c46). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of An Engineered Construct of Phosphatidylinositol 4 Kinase III Beta in Complex with Gtp Gamma S Loaded RAB11, PDB code: 5c46:
In total only one binding site of Magnesium was determined in the Crystal Structure of An Engineered Construct of Phosphatidylinositol 4 Kinase III Beta in Complex with Gtp Gamma S Loaded RAB11, PDB code: 5c46:
Magnesium binding site 1 out of 1 in 5c46
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of An Engineered Construct of Phosphatidylinositol 4 Kinase III Beta in Complex with Gtp Gamma S Loaded RAB11

Mono view
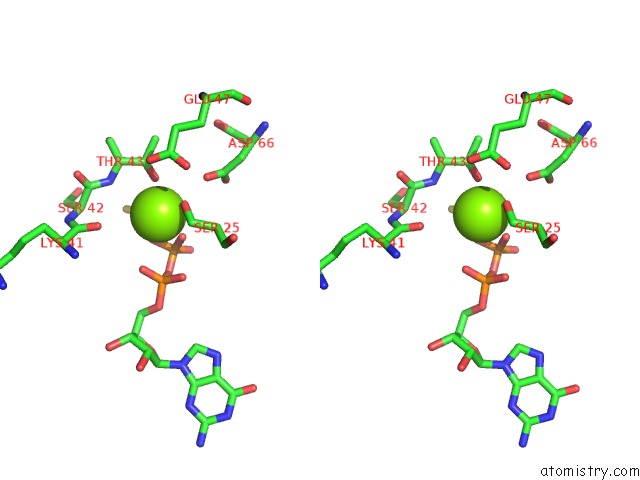
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of An Engineered Construct of Phosphatidylinositol 4 Kinase III Beta in Complex with Gtp Gamma S Loaded RAB11 within 5.0Å range:
|
Reference:
M.L.Fowler,
J.A.Mcphail,
M.L.Jenkins,
G.R.Masson,
F.U.Rutaganira,
K.M.Shokat,
R.L.Williams,
J.E.Burke.
Using Hydrogen Deuterium Exchange Mass Spectrometry to Engineer Optimized Constructs For Crystallization of Protein Complexes: Case Study of PI4KIII Beta with RAB11. Protein Sci. V. 25 826 2016.
ISSN: ESSN 1469-896X
PubMed: 26756197
DOI: 10.1002/PRO.2879
Page generated: Tue Aug 12 06:16:19 2025
ISSN: ESSN 1469-896X
PubMed: 26756197
DOI: 10.1002/PRO.2879
Last articles
Mg in 5QJ7Mg in 5QJ5
Mg in 5QJ6
Mg in 5QJ4
Mg in 5QIN
Mg in 5PRC
Mg in 5PNV
Mg in 5PNW
Mg in 5PNU
Mg in 5PNS