Magnesium »
PDB 5dt3-5e79 »
5e5s »
Magnesium in PDB 5e5s: I-Smami K103A Mutant
Protein crystallography data
The structure of I-Smami K103A Mutant, PDB code: 5e5s
was solved by
B.W.Shen,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 40.00 / 2.29 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 59.589, 68.199, 97.652, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19 / 25.1 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the I-Smami K103A Mutant
(pdb code 5e5s). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the I-Smami K103A Mutant, PDB code: 5e5s:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the I-Smami K103A Mutant, PDB code: 5e5s:
Jump to Magnesium binding site number: 1; 2; 3;
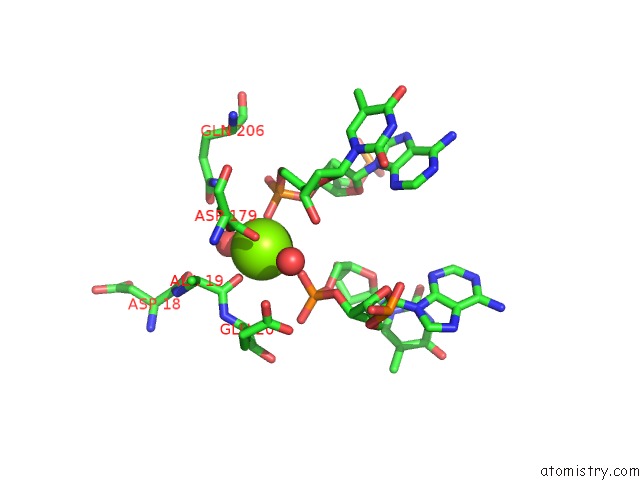
Magnesium binding site 1 out of 3 in 5e5s
Go back to
Magnesium binding site 1 out
of 3 in the I-Smami K103A Mutant
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of I-Smami K103A Mutant within 5.0Å range:
|
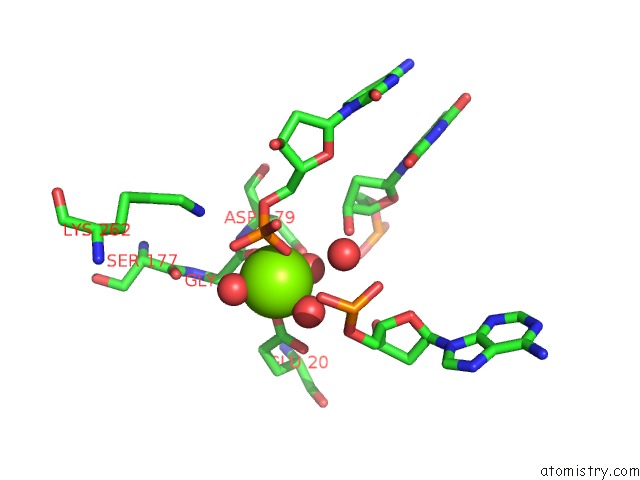
Magnesium binding site 2 out of 3 in 5e5s
Go back to
Magnesium binding site 2 out
of 3 in the I-Smami K103A Mutant
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of I-Smami K103A Mutant within 5.0Å range:
|
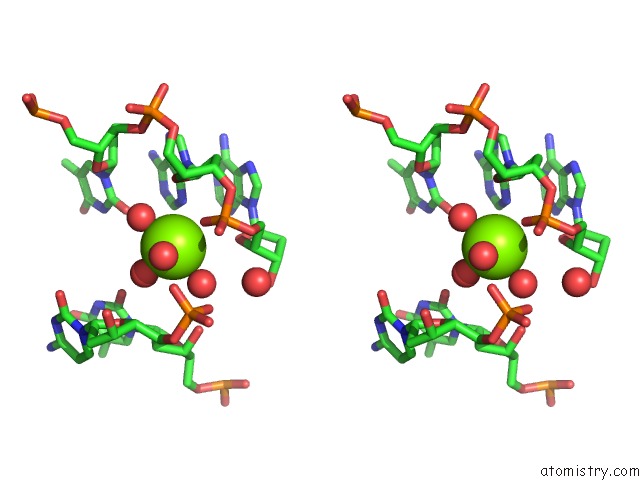
Magnesium binding site 3 out of 3 in 5e5s
Go back to
Magnesium binding site 3 out
of 3 in the I-Smami K103A Mutant

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of I-Smami K103A Mutant within 5.0Å range:
|
Reference:
B.W.Shen,
A.Lambert,
B.C.Walker,
B.L.Stoddard,
B.K.Kaiser.
The Structural Basis of Asymmetry in Dna Binding and Cleavage As Exhibited By the I-Smami Laglidadg Meganuclease. J.Mol.Biol. V. 428 206 2016.
ISSN: ESSN 1089-8638
PubMed: 26705195
DOI: 10.1016/J.JMB.2015.12.005
Page generated: Tue Aug 12 07:31:06 2025
ISSN: ESSN 1089-8638
PubMed: 26705195
DOI: 10.1016/J.JMB.2015.12.005
Last articles
Mg in 5JICMg in 5JDA
Mg in 5JCO
Mg in 5JH7
Mg in 5JD9
Mg in 5JFC
Mg in 5JCB
Mg in 5JCZ
Mg in 5JCP
Mg in 5JCF