Magnesium »
PDB 5fjo-5fuk »
5frm »
Magnesium in PDB 5frm: Crystal Structure of the Prototype Foamy Virus (Pfv) Intasome in Complex with Magnesium and the Insti XZ384 (Compound 4A)
Protein crystallography data
The structure of Crystal Structure of the Prototype Foamy Virus (Pfv) Intasome in Complex with Magnesium and the Insti XZ384 (Compound 4A), PDB code: 5frm
was solved by
D.P.Maskell,
V.E.Pye,
P.Cherepanov,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.90 / 2.58 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 159.737, 159.737, 123.528, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.5 / 20.3 |
Other elements in 5frm:
The structure of Crystal Structure of the Prototype Foamy Virus (Pfv) Intasome in Complex with Magnesium and the Insti XZ384 (Compound 4A) also contains other interesting chemical elements:
| Fluorine | (F) | 2 atoms |
| Zinc | (Zn) | 1 atom |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the Prototype Foamy Virus (Pfv) Intasome in Complex with Magnesium and the Insti XZ384 (Compound 4A)
(pdb code 5frm). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Crystal Structure of the Prototype Foamy Virus (Pfv) Intasome in Complex with Magnesium and the Insti XZ384 (Compound 4A), PDB code: 5frm:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Crystal Structure of the Prototype Foamy Virus (Pfv) Intasome in Complex with Magnesium and the Insti XZ384 (Compound 4A), PDB code: 5frm:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 5frm
Go back to
Magnesium binding site 1 out
of 2 in the Crystal Structure of the Prototype Foamy Virus (Pfv) Intasome in Complex with Magnesium and the Insti XZ384 (Compound 4A)
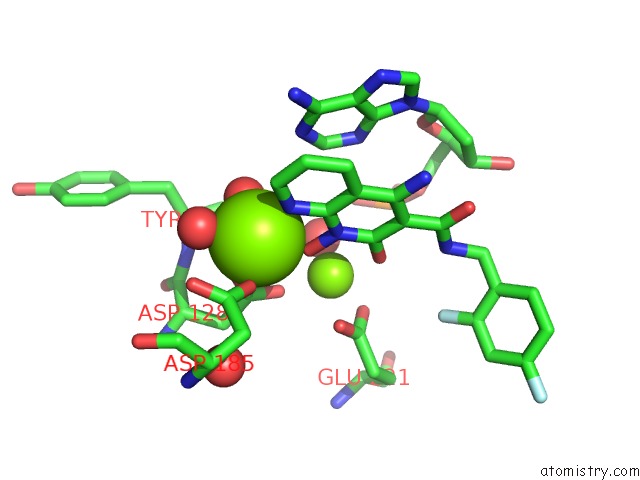
Mono view
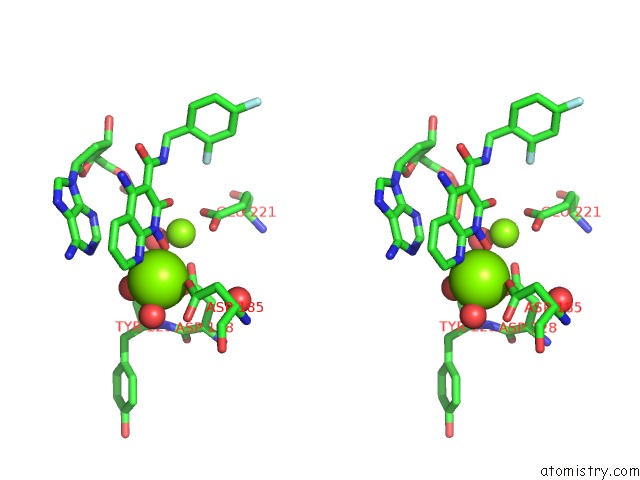
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the Prototype Foamy Virus (Pfv) Intasome in Complex with Magnesium and the Insti XZ384 (Compound 4A) within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 5frm
Go back to
Magnesium binding site 2 out
of 2 in the Crystal Structure of the Prototype Foamy Virus (Pfv) Intasome in Complex with Magnesium and the Insti XZ384 (Compound 4A)
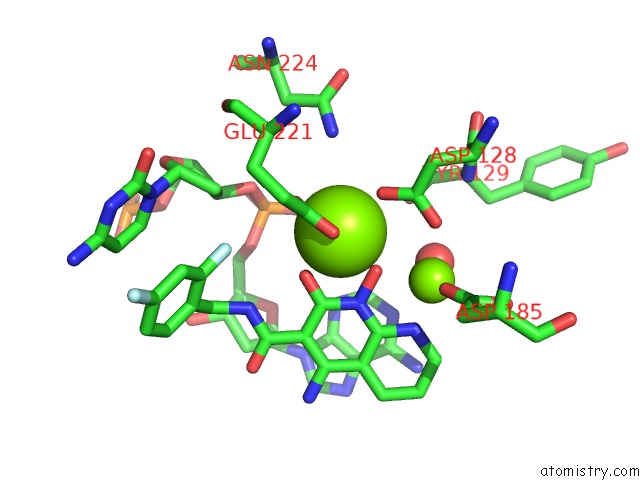
Mono view
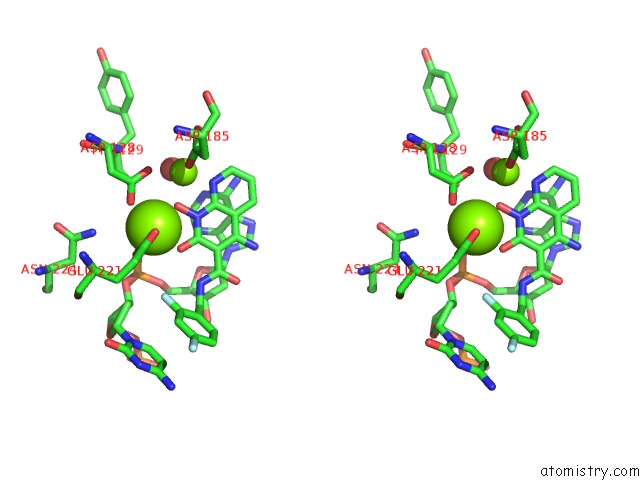
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of the Prototype Foamy Virus (Pfv) Intasome in Complex with Magnesium and the Insti XZ384 (Compound 4A) within 5.0Å range:
|
Reference:
X.Z.Zhao,
S.J.Smith,
D.P.Maskell,
M.Metifiot,
V.E.Pye,
K.Fesen,
C.Marchand,
Y.Pommier,
P.Cherepanov,
S.H.Hughes,
T.R.J.Burke.
Hiv-1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases. Acs Chem.Biol. V. 11 1074 2016.
ISSN: ISSN 1554-8929
PubMed: 26808478
DOI: 10.1021/ACSCHEMBIO.5B00948
Page generated: Sun Sep 29 04:23:37 2024
ISSN: ISSN 1554-8929
PubMed: 26808478
DOI: 10.1021/ACSCHEMBIO.5B00948
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW