Magnesium »
PDB 5fv7-5gg8 »
5g20 »
Magnesium in PDB 5g20: Leishmania Major N-Myristoyltransferase in Complex with A Quinoline Inhibitor (Compound 19).
Enzymatic activity of Leishmania Major N-Myristoyltransferase in Complex with A Quinoline Inhibitor (Compound 19).
All present enzymatic activity of Leishmania Major N-Myristoyltransferase in Complex with A Quinoline Inhibitor (Compound 19).:
2.3.1.97;
2.3.1.97;
Protein crystallography data
The structure of Leishmania Major N-Myristoyltransferase in Complex with A Quinoline Inhibitor (Compound 19)., PDB code: 5g20
was solved by
V.Goncalves,
J.A.Brannigan,
A.Laporte,
A.S.Bell,
S.M.Roberts,
A.J.Wilkinson,
R.J.Leatherbarrow,
E.W.Tate,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.81 / 1.52 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 47.718, 90.651, 52.913, 90.00, 112.72, 90.00 |
| R / Rfree (%) | 17.4 / 21.9 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Leishmania Major N-Myristoyltransferase in Complex with A Quinoline Inhibitor (Compound 19).
(pdb code 5g20). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Leishmania Major N-Myristoyltransferase in Complex with A Quinoline Inhibitor (Compound 19)., PDB code: 5g20:
In total only one binding site of Magnesium was determined in the Leishmania Major N-Myristoyltransferase in Complex with A Quinoline Inhibitor (Compound 19)., PDB code: 5g20:
Magnesium binding site 1 out of 1 in 5g20
Go back to
Magnesium binding site 1 out
of 1 in the Leishmania Major N-Myristoyltransferase in Complex with A Quinoline Inhibitor (Compound 19).
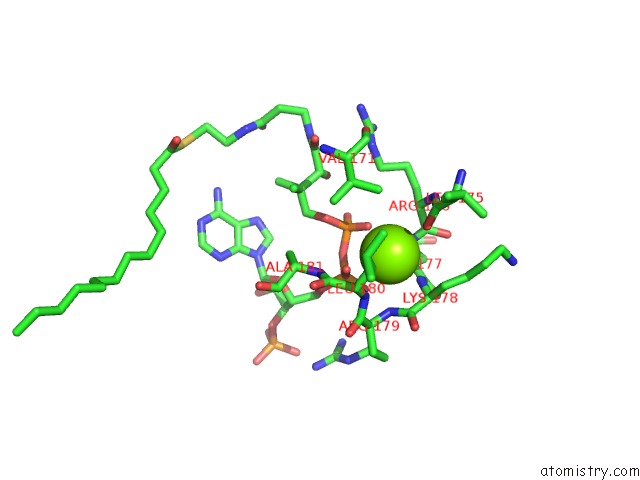
Mono view
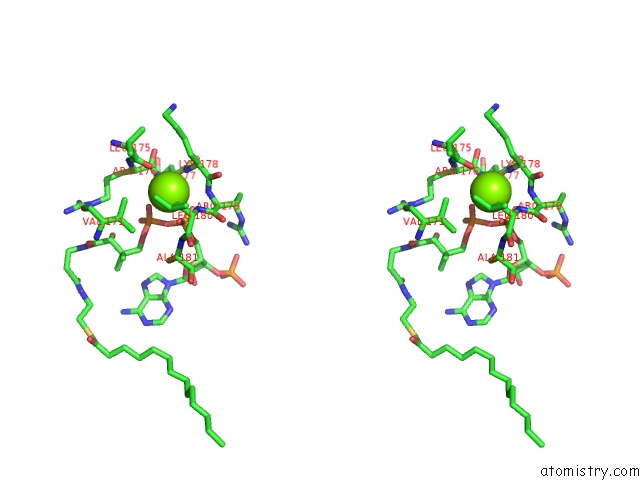
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Leishmania Major N-Myristoyltransferase in Complex with A Quinoline Inhibitor (Compound 19). within 5.0Å range:
|
Reference:
V.Goncalves,
J.A.Brannigan,
A.Laporte,
A.S.Bell,
S.M.Roberts,
A.J.Wilkinson,
R.J.Leatherbarrow,
E.W.Tate.
Structure-Guided Optimization of Quinoline Inhibitors of Plasmodium N-Myristoyltransferase. Medchemcomm V. 8 191 2017.
ISSN: ISSN 2040-2503
PubMed: 28626547
DOI: 10.1039/C6MD00531D
Page generated: Tue Aug 12 08:22:42 2025
ISSN: ISSN 2040-2503
PubMed: 28626547
DOI: 10.1039/C6MD00531D
Last articles
Mg in 6HNQMg in 6HOS
Mg in 6HNS
Mg in 6HN2
Mg in 6HMZ
Mg in 6HMU
Mg in 6HMT
Mg in 6HLR
Mg in 6HLQ
Mg in 6HKY