Magnesium »
PDB 5gg9-5gre »
5gqu »
Magnesium in PDB 5gqu: Crystal Structure of Branching Enzyme From Cyanothece Sp. Atcc 51142
Enzymatic activity of Crystal Structure of Branching Enzyme From Cyanothece Sp. Atcc 51142
All present enzymatic activity of Crystal Structure of Branching Enzyme From Cyanothece Sp. Atcc 51142:
2.4.1.18;
2.4.1.18;
Protein crystallography data
The structure of Crystal Structure of Branching Enzyme From Cyanothece Sp. Atcc 51142, PDB code: 5gqu
was solved by
R.Suzuki,
E.Suzuki,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.29 / 1.85 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 133.750, 133.750, 185.902, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.7 / 17 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Branching Enzyme From Cyanothece Sp. Atcc 51142
(pdb code 5gqu). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Crystal Structure of Branching Enzyme From Cyanothece Sp. Atcc 51142, PDB code: 5gqu:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Crystal Structure of Branching Enzyme From Cyanothece Sp. Atcc 51142, PDB code: 5gqu:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 5gqu
Go back to
Magnesium binding site 1 out
of 4 in the Crystal Structure of Branching Enzyme From Cyanothece Sp. Atcc 51142
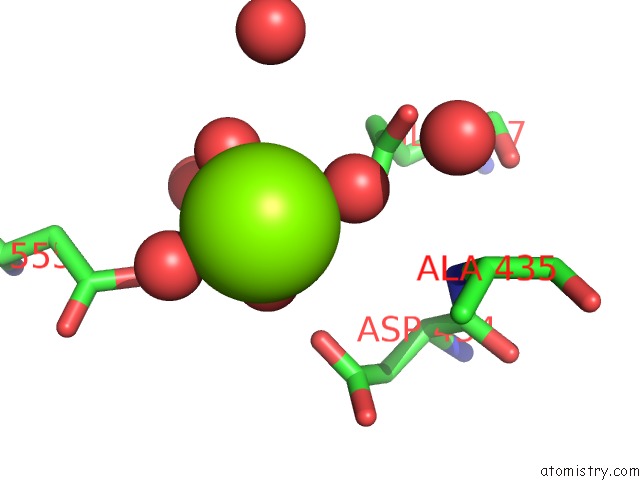
Mono view
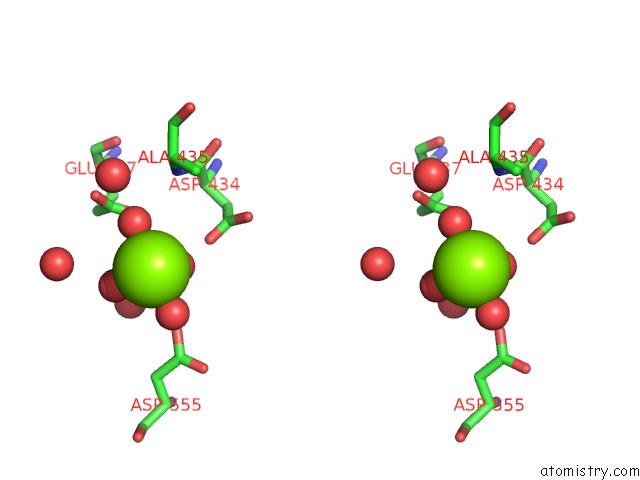
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Branching Enzyme From Cyanothece Sp. Atcc 51142 within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 5gqu
Go back to
Magnesium binding site 2 out
of 4 in the Crystal Structure of Branching Enzyme From Cyanothece Sp. Atcc 51142
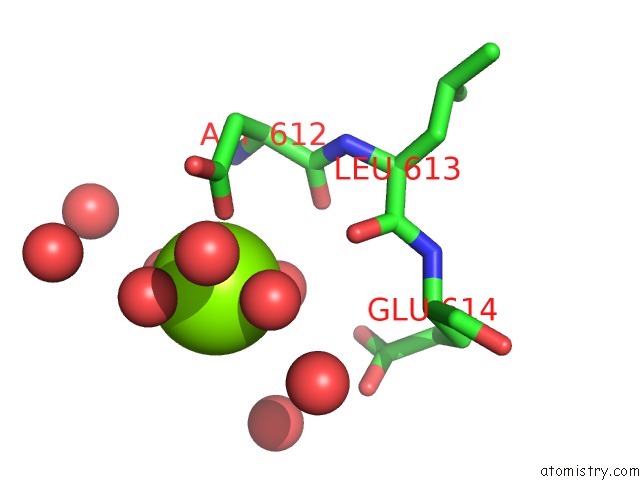
Mono view
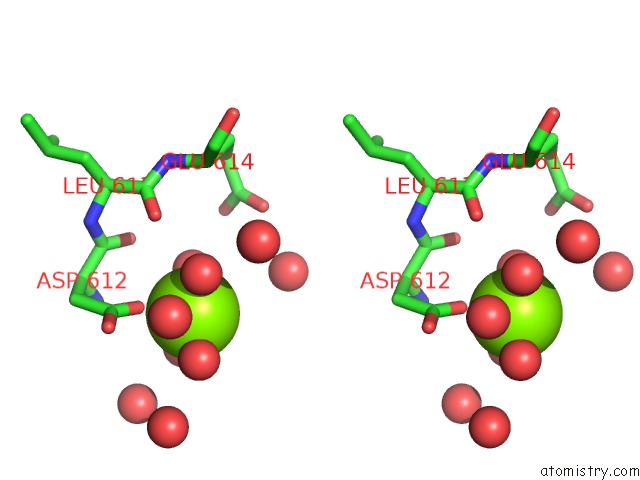
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Branching Enzyme From Cyanothece Sp. Atcc 51142 within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 5gqu
Go back to
Magnesium binding site 3 out
of 4 in the Crystal Structure of Branching Enzyme From Cyanothece Sp. Atcc 51142
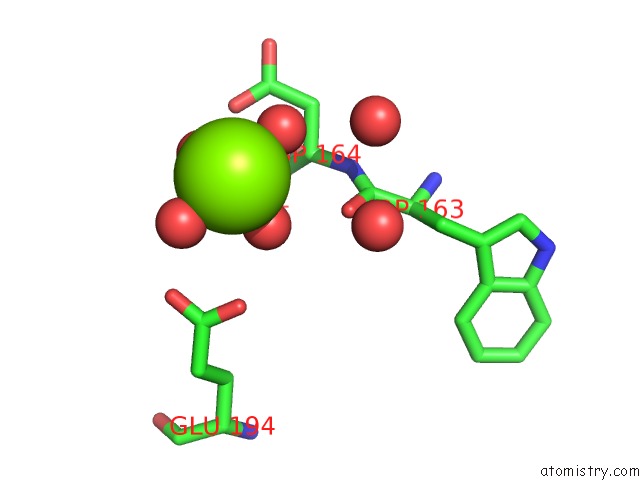
Mono view
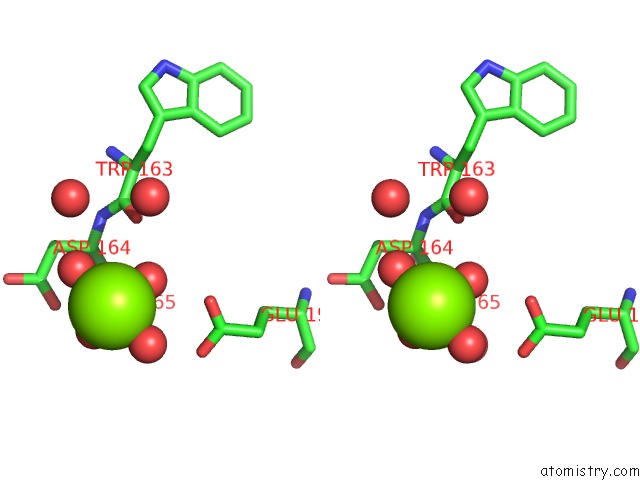
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of Branching Enzyme From Cyanothece Sp. Atcc 51142 within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 5gqu
Go back to
Magnesium binding site 4 out
of 4 in the Crystal Structure of Branching Enzyme From Cyanothece Sp. Atcc 51142
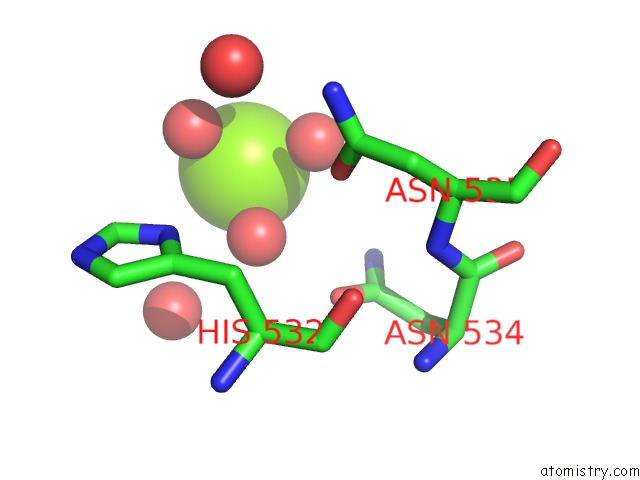
Mono view
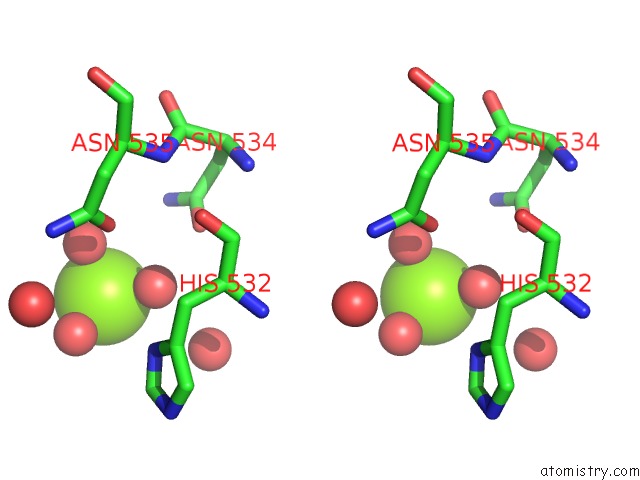
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of Branching Enzyme From Cyanothece Sp. Atcc 51142 within 5.0Å range:
|
Reference:
M.Hayashi,
R.Suzuki,
C.Colleoni,
S.G.Ball,
N.Fujita,
E.Suzuki.
Bound Substrate in the Structure of Cyanobacterial Branching Enzyme Supports A New Mechanistic Model J. Biol. Chem. V. 292 5465 2017.
ISSN: ESSN 1083-351X
PubMed: 28193843
DOI: 10.1074/JBC.M116.755629
Page generated: Tue Aug 12 10:24:06 2025
ISSN: ESSN 1083-351X
PubMed: 28193843
DOI: 10.1074/JBC.M116.755629
Last articles
Mg in 7BODMg in 7BNR
Mg in 7BNK
Mg in 7BMC
Mg in 7BM9
Mg in 7BM8
Mg in 7BM6
Mg in 7BL4
Mg in 7BL6
Mg in 7BL5