Magnesium »
PDB 5ic5-5imp »
5ih0 »
Magnesium in PDB 5ih0: Macrolide 2'-Phosphotransferase Type II Y92M Mutant - Complex with Gdp and Erythromycin
Protein crystallography data
The structure of Macrolide 2'-Phosphotransferase Type II Y92M Mutant - Complex with Gdp and Erythromycin, PDB code: 5ih0
was solved by
A.M.Berghuis,
D.H.Fong,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 41.40 / 1.65 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 39.920, 80.090, 96.710, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.2 / 21 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Macrolide 2'-Phosphotransferase Type II Y92M Mutant - Complex with Gdp and Erythromycin
(pdb code 5ih0). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Macrolide 2'-Phosphotransferase Type II Y92M Mutant - Complex with Gdp and Erythromycin, PDB code: 5ih0:
In total only one binding site of Magnesium was determined in the Macrolide 2'-Phosphotransferase Type II Y92M Mutant - Complex with Gdp and Erythromycin, PDB code: 5ih0:
Magnesium binding site 1 out of 1 in 5ih0
Go back to
Magnesium binding site 1 out
of 1 in the Macrolide 2'-Phosphotransferase Type II Y92M Mutant - Complex with Gdp and Erythromycin
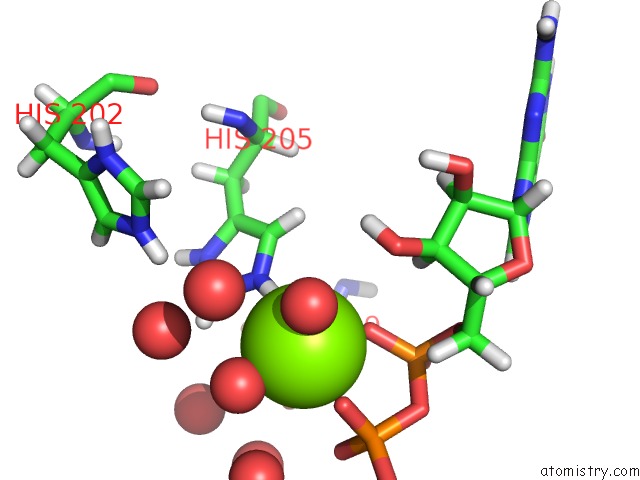
Mono view
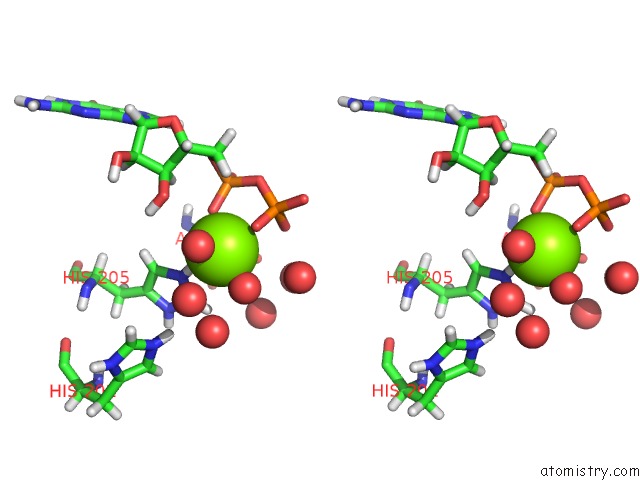
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Macrolide 2'-Phosphotransferase Type II Y92M Mutant - Complex with Gdp and Erythromycin within 5.0Å range:
|
Reference:
D.H.Fong,
D.L.Burk,
J.Blanchet,
A.Y.Yan,
A.M.Berghuis.
Structural Basis For Kinase-Mediated Macrolide Antibiotic Resistance. Structure V. 25 750 2017.
ISSN: ISSN 1878-4186
PubMed: 28416110
DOI: 10.1016/J.STR.2017.03.007
Page generated: Tue Aug 12 11:24:39 2025
ISSN: ISSN 1878-4186
PubMed: 28416110
DOI: 10.1016/J.STR.2017.03.007
Last articles
Mg in 5MF5Mg in 5MCP
Mg in 5MDN
Mg in 5MDL
Mg in 5MDK
Mg in 5MAC
Mg in 5MDJ
Mg in 5MBK
Mg in 5MAQ
Mg in 5MB9