Magnesium »
PDB 5iwx-5j2n »
5ixo »
Magnesium in PDB 5ixo: Crystal Structure of the Arabidopsis Receptor Kinase Haesa Lrr Ectdomain (Apo Form).
Enzymatic activity of Crystal Structure of the Arabidopsis Receptor Kinase Haesa Lrr Ectdomain (Apo Form).
All present enzymatic activity of Crystal Structure of the Arabidopsis Receptor Kinase Haesa Lrr Ectdomain (Apo Form).:
2.7.10.1; 2.7.11.1;
2.7.10.1; 2.7.11.1;
Protein crystallography data
The structure of Crystal Structure of the Arabidopsis Receptor Kinase Haesa Lrr Ectdomain (Apo Form)., PDB code: 5ixo
was solved by
J.Santiago,
M.Hothorn,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 128.76 / 1.74 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 148.678, 148.678, 58.041, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18 / 22.2 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the Arabidopsis Receptor Kinase Haesa Lrr Ectdomain (Apo Form).
(pdb code 5ixo). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of the Arabidopsis Receptor Kinase Haesa Lrr Ectdomain (Apo Form)., PDB code: 5ixo:
In total only one binding site of Magnesium was determined in the Crystal Structure of the Arabidopsis Receptor Kinase Haesa Lrr Ectdomain (Apo Form)., PDB code: 5ixo:
Magnesium binding site 1 out of 1 in 5ixo
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of the Arabidopsis Receptor Kinase Haesa Lrr Ectdomain (Apo Form).

Mono view
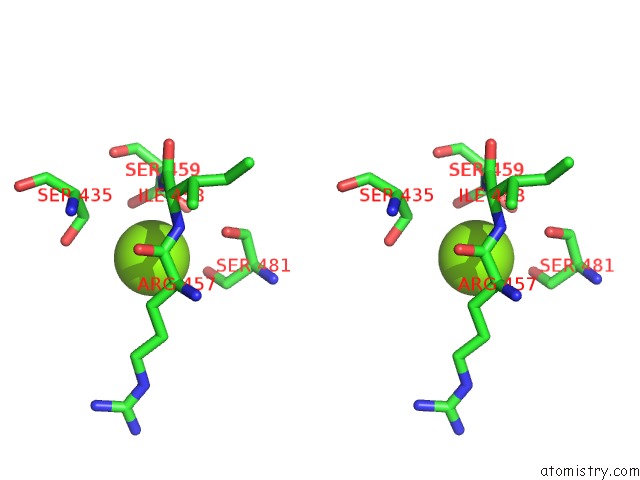
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the Arabidopsis Receptor Kinase Haesa Lrr Ectdomain (Apo Form). within 5.0Å range:
|
Reference:
J.Santiago,
B.Brandt,
M.Wildhagen,
U.Hohmann,
L.A.Hothorn,
M.A.Butenko,
M.Hothorn.
Mechanistic Insight Into A Peptide Hormone Signaling Complex Mediating Floral Organ Abscission. Elife V. 5 2016.
ISSN: ESSN 2050-084X
PubMed: 27058169
DOI: 10.7554/ELIFE.15075
Page generated: Tue Aug 12 11:55:10 2025
ISSN: ESSN 2050-084X
PubMed: 27058169
DOI: 10.7554/ELIFE.15075
Last articles
Mg in 6QPHMg in 6QV0
Mg in 6QUZ
Mg in 6QUY
Mg in 6QUX
Mg in 6QUW
Mg in 6QUV
Mg in 6QUU
Mg in 6QUS
Mg in 6QTN