Magnesium »
PDB 5lu5-5m5i »
5m1t »
Magnesium in PDB 5m1t: Pamucr Phosphodiesterase, C-Di-Gmp Complex
Protein crystallography data
The structure of Pamucr Phosphodiesterase, C-Di-Gmp Complex, PDB code: 5m1t
was solved by
A.Hutchin,
I.Tews,
M.A.Walsh,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.27 / 2.27 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 46.370, 116.110, 52.140, 90.00, 102.52, 90.00 |
| R / Rfree (%) | 19.5 / 23.8 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Pamucr Phosphodiesterase, C-Di-Gmp Complex
(pdb code 5m1t). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Pamucr Phosphodiesterase, C-Di-Gmp Complex, PDB code: 5m1t:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Pamucr Phosphodiesterase, C-Di-Gmp Complex, PDB code: 5m1t:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 5m1t
Go back to
Magnesium binding site 1 out
of 4 in the Pamucr Phosphodiesterase, C-Di-Gmp Complex
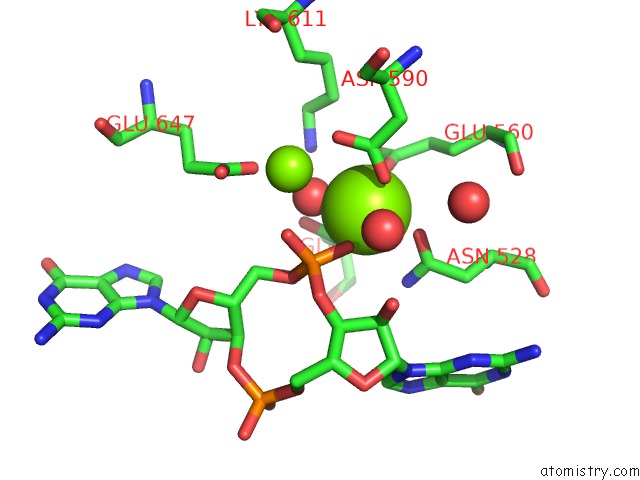
Mono view
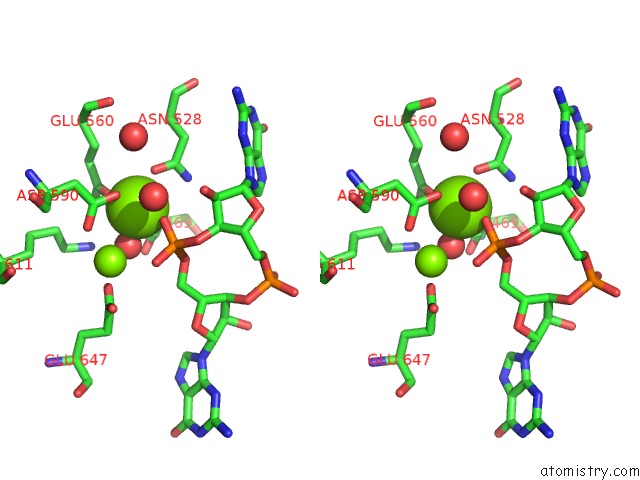
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Pamucr Phosphodiesterase, C-Di-Gmp Complex within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 5m1t
Go back to
Magnesium binding site 2 out
of 4 in the Pamucr Phosphodiesterase, C-Di-Gmp Complex
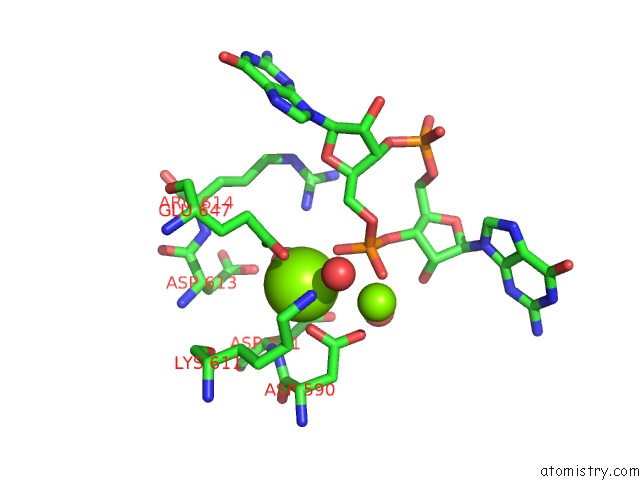
Mono view
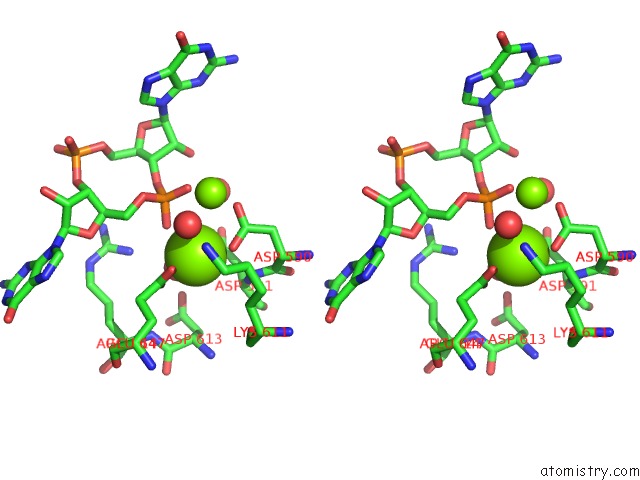
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Pamucr Phosphodiesterase, C-Di-Gmp Complex within 5.0Å range:
|
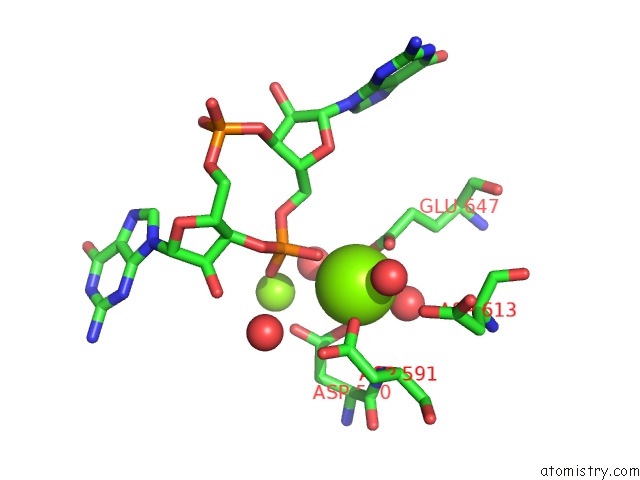
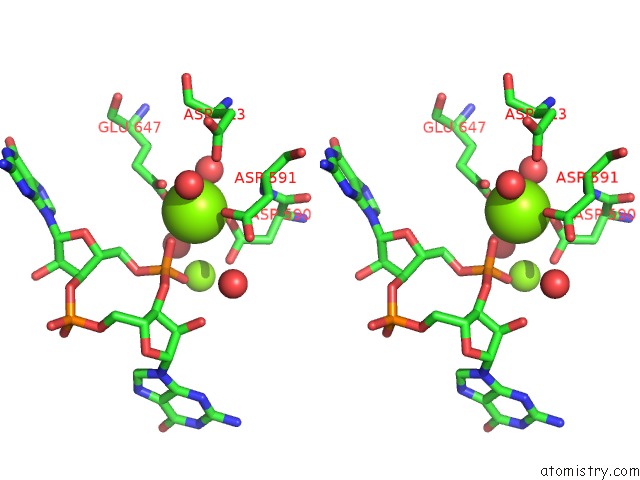
Magnesium binding site 3 out of 4 in 5m1t
Go back to
Magnesium binding site 3 out
of 4 in the Pamucr Phosphodiesterase, C-Di-Gmp Complex
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Pamucr Phosphodiesterase, C-Di-Gmp Complex within 5.0Å range:
|
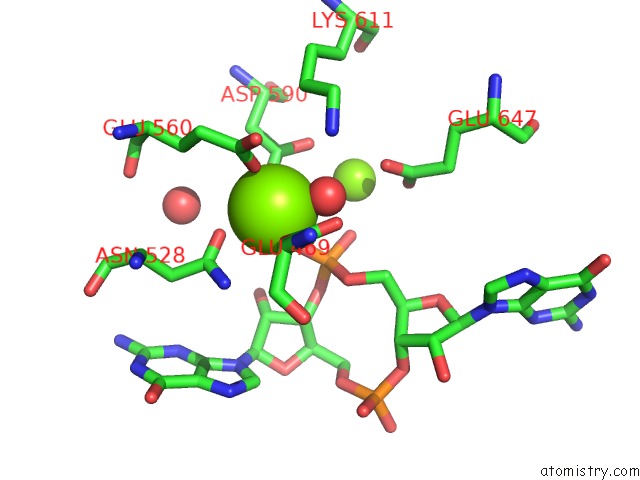
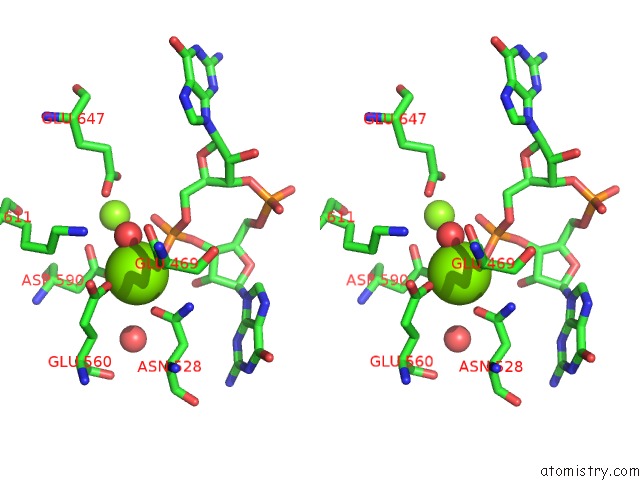
Magnesium binding site 4 out of 4 in 5m1t
Go back to
Magnesium binding site 4 out
of 4 in the Pamucr Phosphodiesterase, C-Di-Gmp Complex
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Pamucr Phosphodiesterase, C-Di-Gmp Complex within 5.0Å range:
|
Reference:
D.Bellini,
S.Horrell,
A.Hutchin,
C.W.Phippen,
R.W.Strange,
Y.Cai,
A.Wagner,
J.S.Webb,
I.Tews,
M.A.Walsh.
Dimerisation Induced Formation of the Active Site and the Identification of Three Metal Sites in Eal-Phosphodiesterases. Sci Rep V. 7 42166 2017.
ISSN: ESSN 2045-2322
PubMed: 28186120
DOI: 10.1038/SREP42166
Page generated: Sun Sep 29 21:08:34 2024
ISSN: ESSN 2045-2322
PubMed: 28186120
DOI: 10.1038/SREP42166
Last articles
Ca in 5UUYCa in 5UUH
Ca in 5UUG
Ca in 5UUD
Ca in 5UUC
Ca in 5UUB
Ca in 5UUA
Ca in 5UU9
Ca in 5UU8
Ca in 5UN2