Magnesium »
PDB 5lu5-5m5i »
5m3f »
Magnesium in PDB 5m3f: Yeast Rna Polymerase I Elongation Complex at 3.8A
Enzymatic activity of Yeast Rna Polymerase I Elongation Complex at 3.8A
All present enzymatic activity of Yeast Rna Polymerase I Elongation Complex at 3.8A:
2.7.7.6;
2.7.7.6;
Other elements in 5m3f:
The structure of Yeast Rna Polymerase I Elongation Complex at 3.8A also contains other interesting chemical elements:
| Zinc | (Zn) | 6 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Yeast Rna Polymerase I Elongation Complex at 3.8A
(pdb code 5m3f). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Yeast Rna Polymerase I Elongation Complex at 3.8A, PDB code: 5m3f:
In total only one binding site of Magnesium was determined in the Yeast Rna Polymerase I Elongation Complex at 3.8A, PDB code: 5m3f:
Magnesium binding site 1 out of 1 in 5m3f
Go back to
Magnesium binding site 1 out
of 1 in the Yeast Rna Polymerase I Elongation Complex at 3.8A
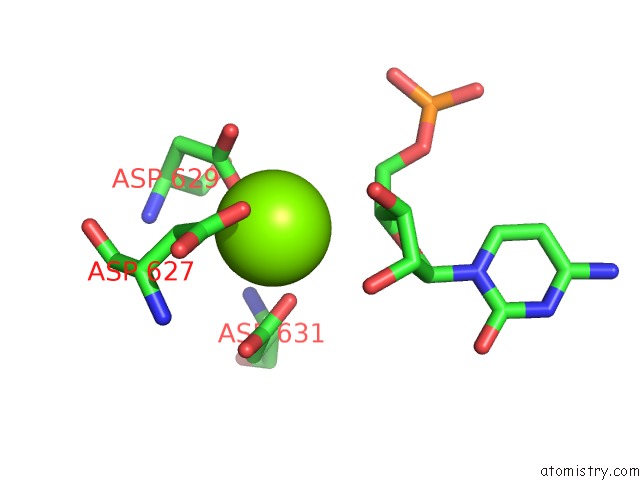
Mono view
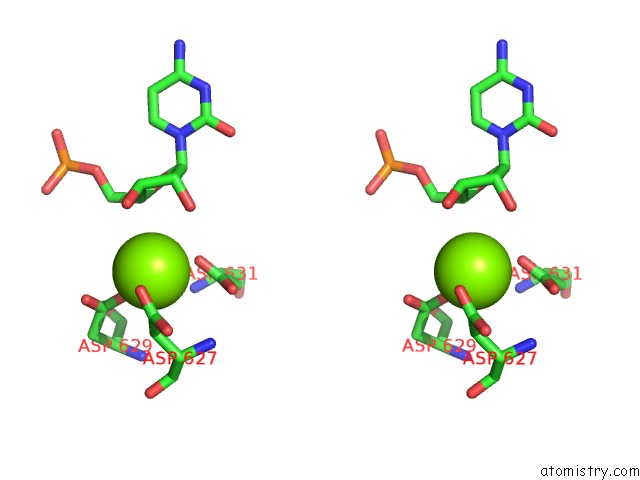
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Yeast Rna Polymerase I Elongation Complex at 3.8A within 5.0Å range:
|
Reference:
S.Neyer,
M.Kunz,
C.Geiss,
M.Hantsche,
V.V.Hodirnau,
A.Seybert,
C.Engel,
M.P.Scheffer,
P.Cramer,
A.S.Frangakis.
Structure of Rna Polymerase I Transcribing Ribosomal Dna Genes. Nature V. 540 607 2016.
ISSN: ISSN 0028-0836
PubMed: 27842382
DOI: 10.1038/NATURE20561
Page generated: Sun Sep 29 21:09:37 2024
ISSN: ISSN 0028-0836
PubMed: 27842382
DOI: 10.1038/NATURE20561
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO