Magnesium »
PDB 5lu5-5m5i »
5m5c »
Magnesium in PDB 5m5c: Mechanism of Microtubule Minus-End Recognition and Protection By Camsap Proteins
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Mechanism of Microtubule Minus-End Recognition and Protection By Camsap Proteins
(pdb code 5m5c). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Mechanism of Microtubule Minus-End Recognition and Protection By Camsap Proteins, PDB code: 5m5c:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Mechanism of Microtubule Minus-End Recognition and Protection By Camsap Proteins, PDB code: 5m5c:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 5m5c
Go back to
Magnesium binding site 1 out
of 2 in the Mechanism of Microtubule Minus-End Recognition and Protection By Camsap Proteins
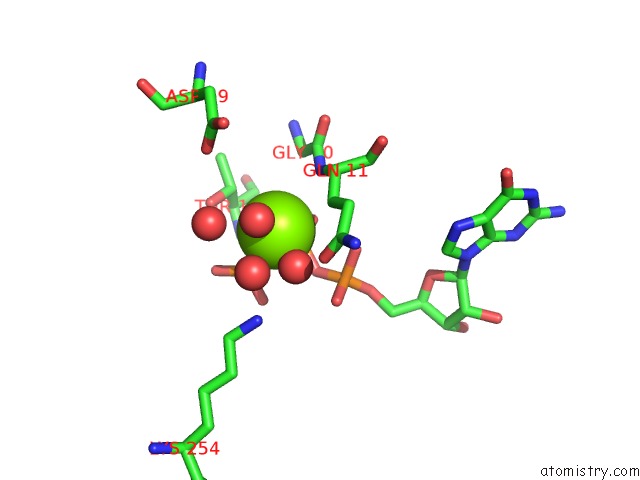
Mono view
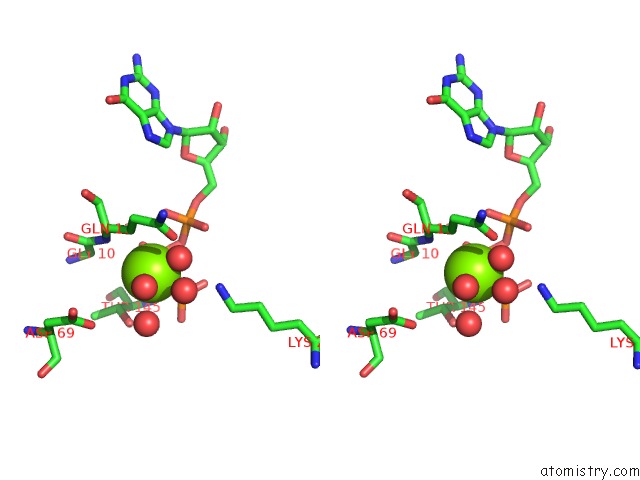
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Mechanism of Microtubule Minus-End Recognition and Protection By Camsap Proteins within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 5m5c
Go back to
Magnesium binding site 2 out
of 2 in the Mechanism of Microtubule Minus-End Recognition and Protection By Camsap Proteins
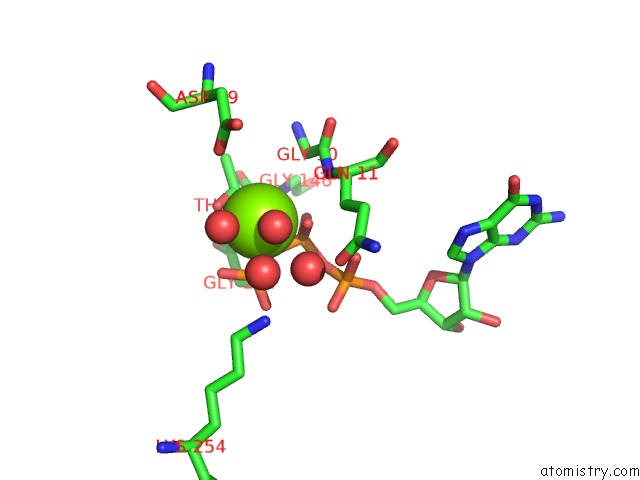
Mono view
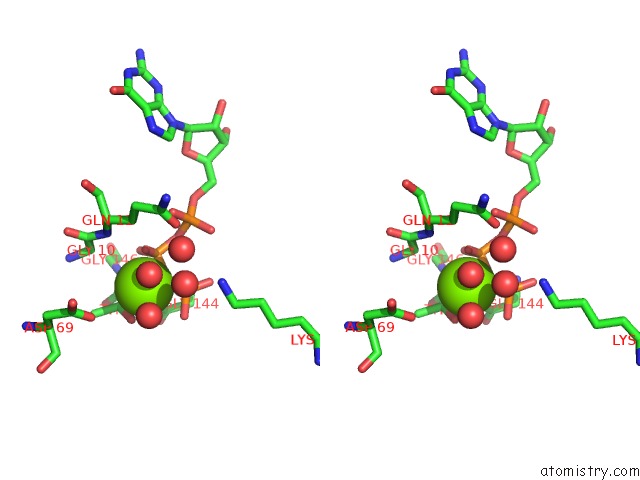
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Mechanism of Microtubule Minus-End Recognition and Protection By Camsap Proteins within 5.0Å range:
|
Reference:
J.Atherton,
K.Jiang,
M.M.Stangier,
Y.Luo,
S.Hua,
K.Houben,
J.J.E.Van Hooff,
A.P.Joseph,
G.Scarabelli,
B.J.Grant,
A.J.Roberts,
M.Topf,
M.O.Steinmetz,
M.Baldus,
C.A.Moores,
A.Akhmanova.
A Structural Model For Microtubule Minus-End Recognition and Protection By Camsap Proteins. Nat. Struct. Mol. Biol. V. 24 931 2017.
ISSN: ESSN 1545-9985
PubMed: 28991265
DOI: 10.1038/NSMB.3483
Page generated: Sun Sep 29 21:10:55 2024
ISSN: ESSN 1545-9985
PubMed: 28991265
DOI: 10.1038/NSMB.3483
Last articles
Cl in 7THHCl in 7TIV
Cl in 7TIW
Cl in 7TI9
Cl in 7TIU
Cl in 7TIN
Cl in 7TI7
Cl in 7TI1
Cl in 7TI0
Cl in 7TH2