Magnesium »
PDB 5m5l-5mh5 »
5m70 »
Magnesium in PDB 5m70: Crystal Structure of Human Rhogap Mutated in Its Arginin Finger (R85A) in Complex with Rhoa.Gdp.ALF4- Human
Protein crystallography data
The structure of Crystal Structure of Human Rhogap Mutated in Its Arginin Finger (R85A) in Complex with Rhoa.Gdp.ALF4- Human, PDB code: 5m70
was solved by
E.Pellegrini,
M.W.Bowler,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 2.20 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 72.554, 66.063, 76.759, 90.00, 96.07, 90.00 |
| R / Rfree (%) | 23.8 / 27.7 |
Other elements in 5m70:
The structure of Crystal Structure of Human Rhogap Mutated in Its Arginin Finger (R85A) in Complex with Rhoa.Gdp.ALF4- Human also contains other interesting chemical elements:
| Fluorine | (F) | 8 atoms |
| Aluminium | (Al) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Human Rhogap Mutated in Its Arginin Finger (R85A) in Complex with Rhoa.Gdp.ALF4- Human
(pdb code 5m70). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Crystal Structure of Human Rhogap Mutated in Its Arginin Finger (R85A) in Complex with Rhoa.Gdp.ALF4- Human, PDB code: 5m70:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Crystal Structure of Human Rhogap Mutated in Its Arginin Finger (R85A) in Complex with Rhoa.Gdp.ALF4- Human, PDB code: 5m70:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 5m70
Go back to
Magnesium binding site 1 out
of 2 in the Crystal Structure of Human Rhogap Mutated in Its Arginin Finger (R85A) in Complex with Rhoa.Gdp.ALF4- Human
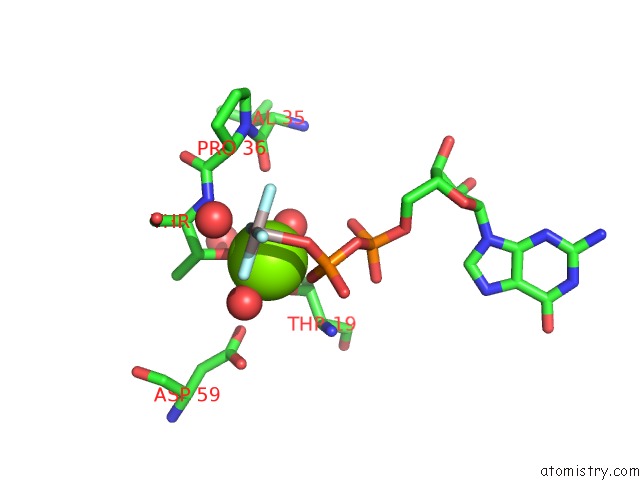
Mono view
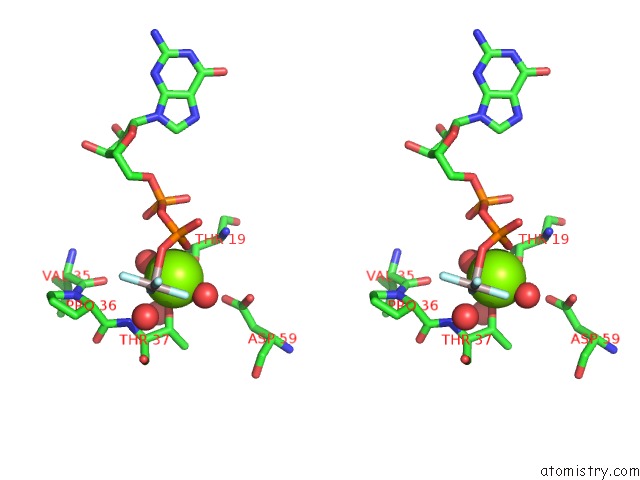
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Human Rhogap Mutated in Its Arginin Finger (R85A) in Complex with Rhoa.Gdp.ALF4- Human within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 5m70
Go back to
Magnesium binding site 2 out
of 2 in the Crystal Structure of Human Rhogap Mutated in Its Arginin Finger (R85A) in Complex with Rhoa.Gdp.ALF4- Human
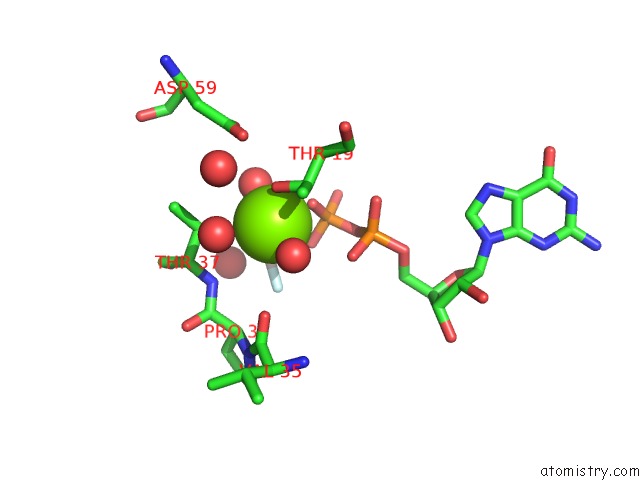
Mono view
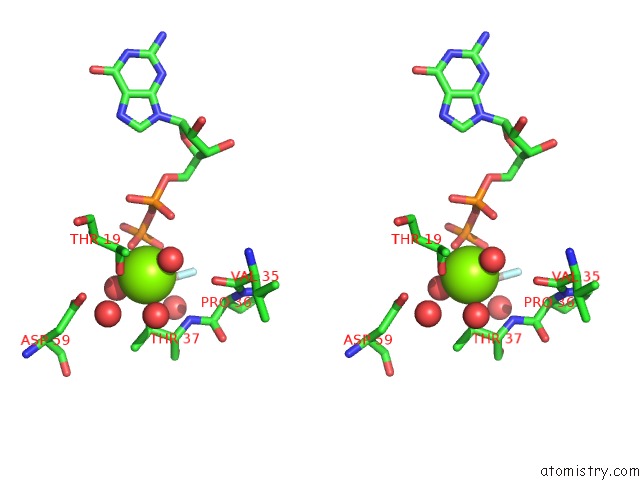
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Human Rhogap Mutated in Its Arginin Finger (R85A) in Complex with Rhoa.Gdp.ALF4- Human within 5.0Å range:
|
Reference:
Y.Jin,
R.W.Molt,
E.Pellegrini,
M.J.Cliff,
M.W.Bowler,
N.G.J.Richards,
G.M.Blackburn,
J.P.Waltho.
Assessing the Influence of Mutation on Gtpase Transition States By Using X-Ray Crystallography, (19) F uc(Nmr), and Dft Approaches. Angew. Chem. Int. Ed. Engl. V. 56 9732 2017.
ISSN: ESSN 1521-3773
PubMed: 28498638
DOI: 10.1002/ANIE.201703074
Page generated: Sun Sep 29 21:14:42 2024
ISSN: ESSN 1521-3773
PubMed: 28498638
DOI: 10.1002/ANIE.201703074
Last articles
Cl in 6BMZCl in 6BN2
Cl in 6BN0
Cl in 6BMW
Cl in 6BMU
Cl in 6BMX
Cl in 6BMR
Cl in 6BMV
Cl in 6BMT
Cl in 6BLA