Magnesium »
PDB 6b5e-6bcr »
6b6x »
Magnesium in PDB 6b6x: Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster
Enzymatic activity of Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster
All present enzymatic activity of Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster:
1.2.7.4;
1.2.7.4;
Protein crystallography data
The structure of Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster, PDB code: 6b6x
was solved by
E.C.Wittenborn,
C.L.Drennan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 64.87 / 1.84 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 64.650, 154.870, 66.490, 90.00, 102.67, 90.00 |
| R / Rfree (%) | 14.4 / 17.1 |
Other elements in 6b6x:
The structure of Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster also contains other interesting chemical elements:
| Nickel | (Ni) | 2 atoms |
| Iron | (Fe) | 18 atoms |
| Chlorine | (Cl) | 3 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster
(pdb code 6b6x). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster, PDB code: 6b6x:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster, PDB code: 6b6x:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 6b6x
Go back to
Magnesium binding site 1 out
of 4 in the Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster
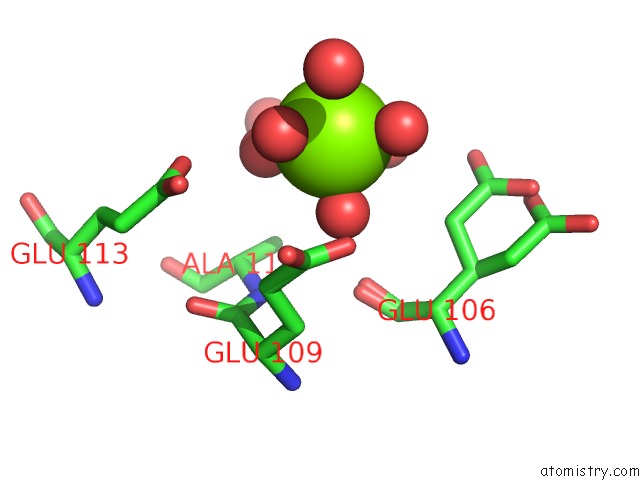
Mono view
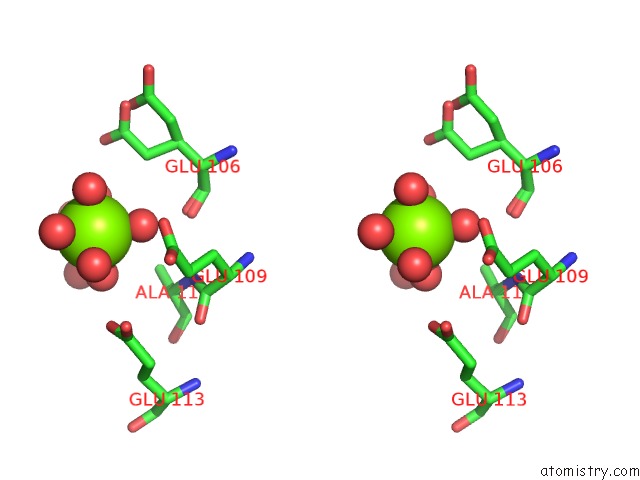
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 6b6x
Go back to
Magnesium binding site 2 out
of 4 in the Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster
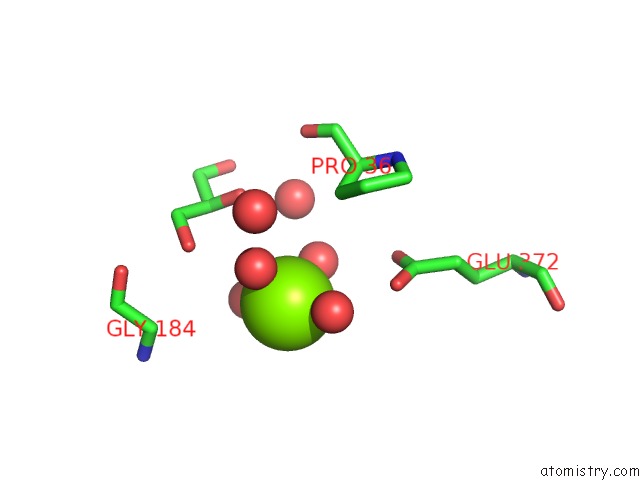
Mono view
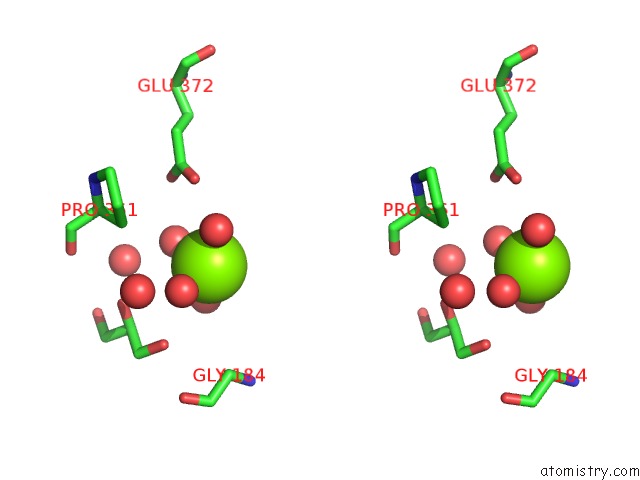
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 6b6x
Go back to
Magnesium binding site 3 out
of 4 in the Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 6b6x
Go back to
Magnesium binding site 4 out
of 4 in the Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C- Cluster within 5.0Å range:
|
Reference:
E.C.Wittenborn,
M.Merrouch,
C.Ueda,
L.Fradale,
C.Leger,
V.Fourmond,
M.E.Pandelia,
S.Dementin,
C.L.Drennan.
Redox-Dependent Rearrangements of the Nifes Cluster of Carbon Monoxide Dehydrogenase. Elife V. 7 2018.
ISSN: ESSN 2050-084X
PubMed: 30277213
DOI: 10.7554/ELIFE.39451
Page generated: Mon Sep 30 19:37:09 2024
ISSN: ESSN 2050-084X
PubMed: 30277213
DOI: 10.7554/ELIFE.39451
Last articles
F in 4G4MF in 4GJ3
F in 4GHT
F in 4GJ2
F in 4GHI
F in 4GG7
F in 4GG5
F in 4GE6
F in 4GE5
F in 4GCA