Magnesium »
PDB 6cgf-6cq2 »
6cj3 »
Magnesium in PDB 6cj3: Crystal Structure of Protein Cite From Mycobacterium Tuberculosis in Complex with Magnesium and Pyruvate
Protein crystallography data
The structure of Crystal Structure of Protein Cite From Mycobacterium Tuberculosis in Complex with Magnesium and Pyruvate, PDB code: 6cj3
was solved by
E.V.Fedorov,
A.A.Fedorov,
H.Wang,
J.B.Bonanno,
L.Carvalho,
S.C.Almo,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.00 / 1.73 |
| Space group | H 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 91.992, 91.992, 221.137, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 19.3 / 21.8 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Protein Cite From Mycobacterium Tuberculosis in Complex with Magnesium and Pyruvate
(pdb code 6cj3). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of Protein Cite From Mycobacterium Tuberculosis in Complex with Magnesium and Pyruvate, PDB code: 6cj3:
In total only one binding site of Magnesium was determined in the Crystal Structure of Protein Cite From Mycobacterium Tuberculosis in Complex with Magnesium and Pyruvate, PDB code: 6cj3:
Magnesium binding site 1 out of 1 in 6cj3
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of Protein Cite From Mycobacterium Tuberculosis in Complex with Magnesium and Pyruvate
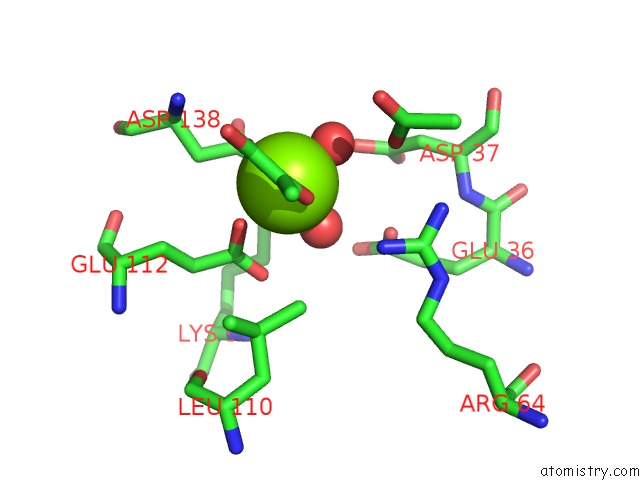
Mono view
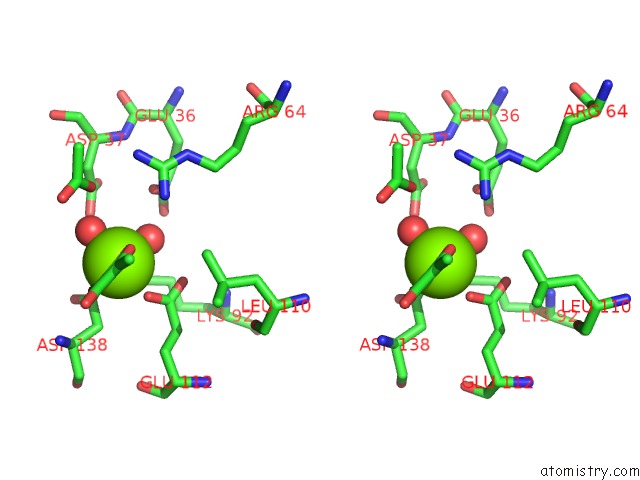
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Protein Cite From Mycobacterium Tuberculosis in Complex with Magnesium and Pyruvate within 5.0Å range:
|
Reference:
H.Wang,
A.A.Fedorov,
E.V.Fedorov,
D.M.Hunt,
A.Rodgers,
H.L.Douglas,
A.Garza-Garcia,
J.B.Bonanno,
S.C.Almo,
L.P.S.De Carvalho.
An Essential Bifunctional Enzyme Inmycobacterium Tuberculosisfor Itaconate Dissimilation and Leucine Catabolism. Proc.Natl.Acad.Sci.Usa V. 116 15907 2019.
ISSN: ESSN 1091-6490
PubMed: 31320588
DOI: 10.1073/PNAS.1906606116
Page generated: Wed Aug 13 04:36:06 2025
ISSN: ESSN 1091-6490
PubMed: 31320588
DOI: 10.1073/PNAS.1906606116
Last articles
Mg in 6YRJMg in 6YRD
Mg in 6YR8
Mg in 6YQ4
Mg in 6YR4
Mg in 6YPN
Mg in 6YOM
Mg in 6YL5
Mg in 6YP4
Mg in 6YOX