Magnesium »
PDB 6d9s-6dmd »
6djo »
Magnesium in PDB 6djo: Cryo-Em Structure of Adp-Actin Filaments
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of Adp-Actin Filaments
(pdb code 6djo). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Cryo-Em Structure of Adp-Actin Filaments, PDB code: 6djo:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Cryo-Em Structure of Adp-Actin Filaments, PDB code: 6djo:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 6djo
Go back to
Magnesium binding site 1 out
of 4 in the Cryo-Em Structure of Adp-Actin Filaments
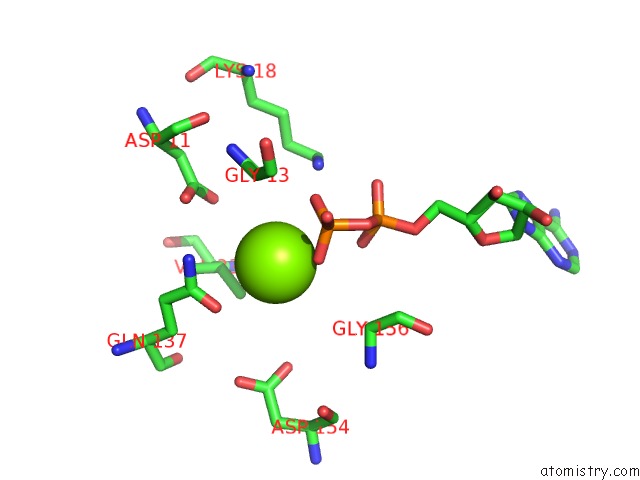
Mono view
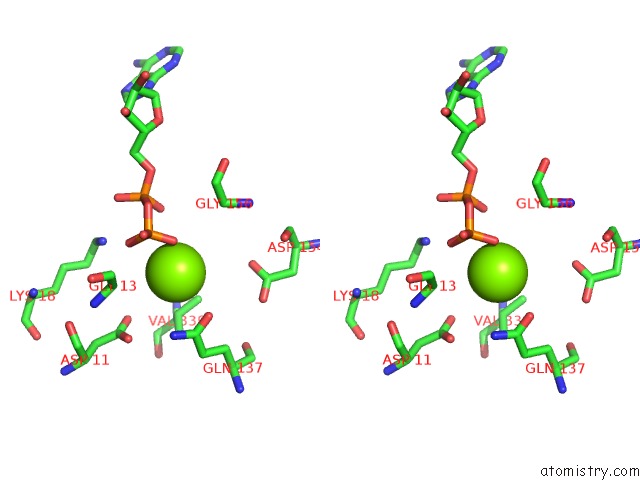
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of Adp-Actin Filaments within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 6djo
Go back to
Magnesium binding site 2 out
of 4 in the Cryo-Em Structure of Adp-Actin Filaments
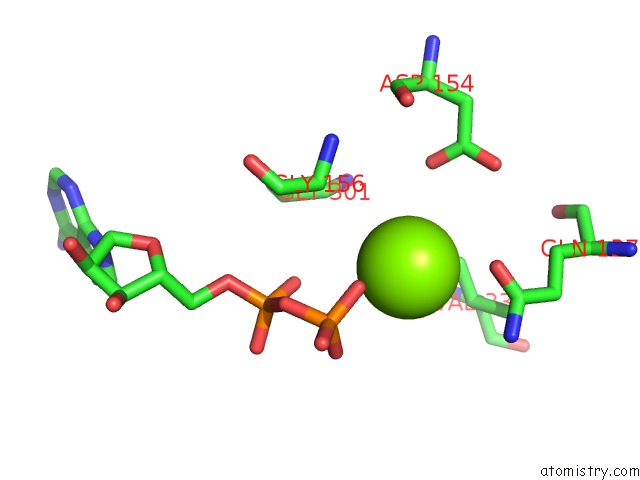
Mono view
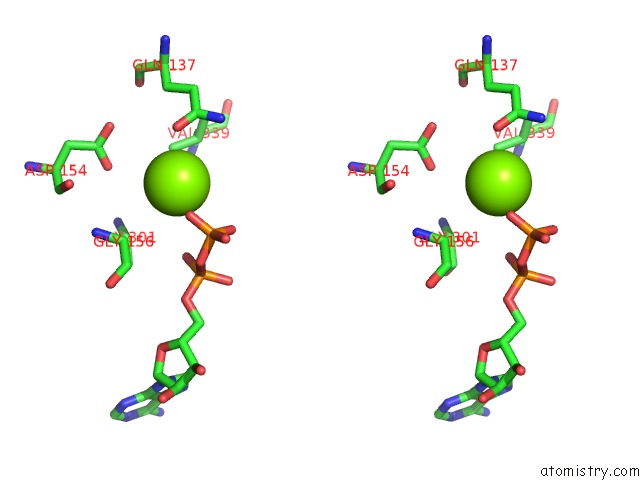
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Cryo-Em Structure of Adp-Actin Filaments within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 6djo
Go back to
Magnesium binding site 3 out
of 4 in the Cryo-Em Structure of Adp-Actin Filaments
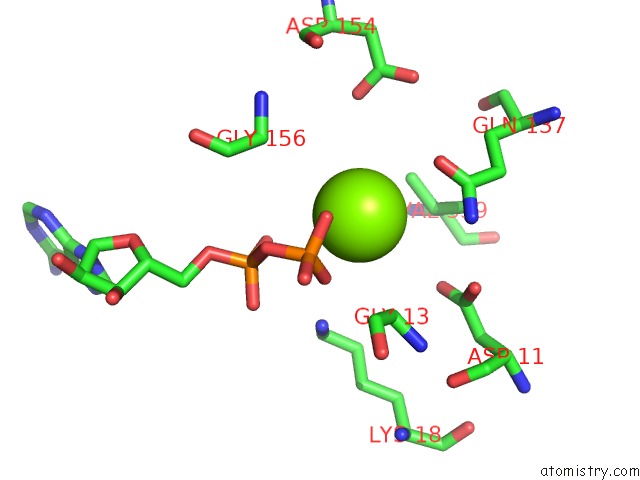
Mono view
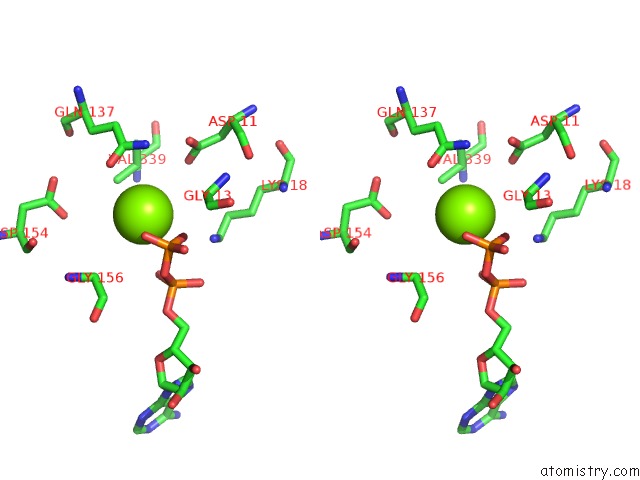
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Cryo-Em Structure of Adp-Actin Filaments within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 6djo
Go back to
Magnesium binding site 4 out
of 4 in the Cryo-Em Structure of Adp-Actin Filaments
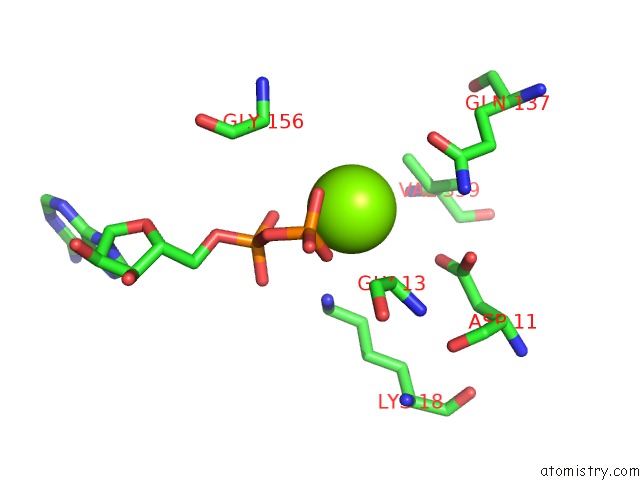
Mono view
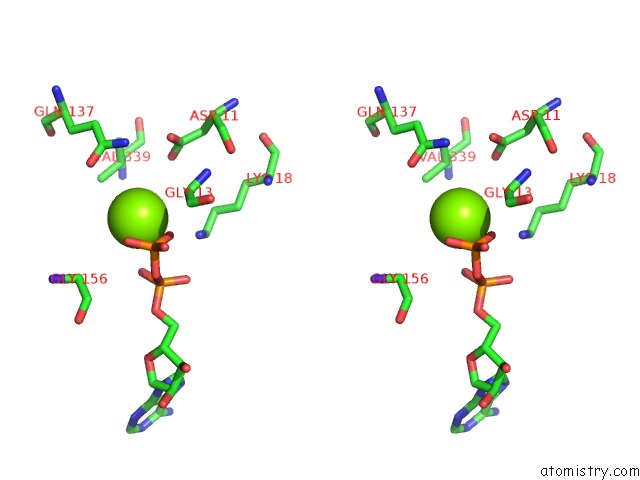
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Cryo-Em Structure of Adp-Actin Filaments within 5.0Å range:
|
Reference:
S.Z.Chou,
T.D.Pollard.
Mechanism of Actin Polymerization Revealed By Cryo-Em Structures of Actin Filaments with Three Different Bound Nucleotides. Proc.Natl.Acad.Sci.Usa V. 116 4265 2019.
ISSN: ESSN 1091-6490
PubMed: 30760599
DOI: 10.1073/PNAS.1807028115
Page generated: Wed Aug 13 05:08:29 2025
ISSN: ESSN 1091-6490
PubMed: 30760599
DOI: 10.1073/PNAS.1807028115
Last articles
Mg in 6YRCMg in 6YRJ
Mg in 6YRD
Mg in 6YR8
Mg in 6YQ4
Mg in 6YR4
Mg in 6YPN
Mg in 6YOM
Mg in 6YL5
Mg in 6YP4