Magnesium »
PDB 6e8u-6el4 »
6ehh »
Magnesium in PDB 6ehh: Crystal Structure of Mouse MTH1 Mutant L116M with Inhibitor TH588
Enzymatic activity of Crystal Structure of Mouse MTH1 Mutant L116M with Inhibitor TH588
All present enzymatic activity of Crystal Structure of Mouse MTH1 Mutant L116M with Inhibitor TH588:
3.6.1.55; 3.6.1.56;
3.6.1.55; 3.6.1.56;
Protein crystallography data
The structure of Crystal Structure of Mouse MTH1 Mutant L116M with Inhibitor TH588, PDB code: 6ehh
was solved by
R.Gustafsson,
M.Narwal,
A.-S.Jemth,
I.Almlof,
U.Warpman Berglund,
T.Helleday,
P.Stenmark,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.40 / 2.40 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.673, 142.088, 58.750, 90.00, 107.52, 90.00 |
| R / Rfree (%) | 24.9 / 29.9 |
Other elements in 6ehh:
The structure of Crystal Structure of Mouse MTH1 Mutant L116M with Inhibitor TH588 also contains other interesting chemical elements:
| Copper | (Cu) | 4 atoms |
| Chlorine | (Cl) | 8 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Mouse MTH1 Mutant L116M with Inhibitor TH588
(pdb code 6ehh). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of Mouse MTH1 Mutant L116M with Inhibitor TH588, PDB code: 6ehh:
In total only one binding site of Magnesium was determined in the Crystal Structure of Mouse MTH1 Mutant L116M with Inhibitor TH588, PDB code: 6ehh:
Magnesium binding site 1 out of 1 in 6ehh
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of Mouse MTH1 Mutant L116M with Inhibitor TH588
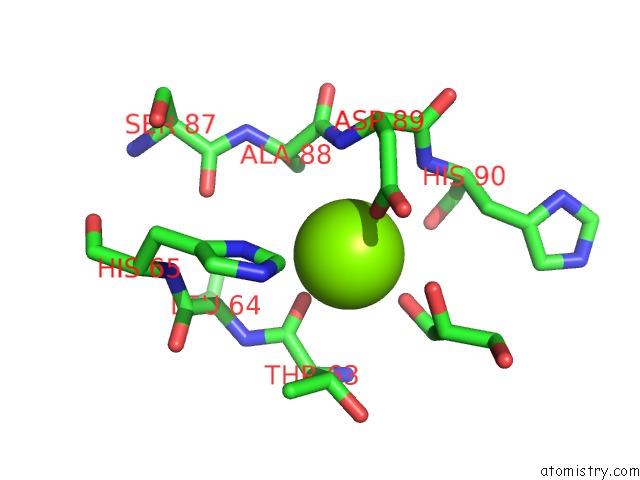
Mono view
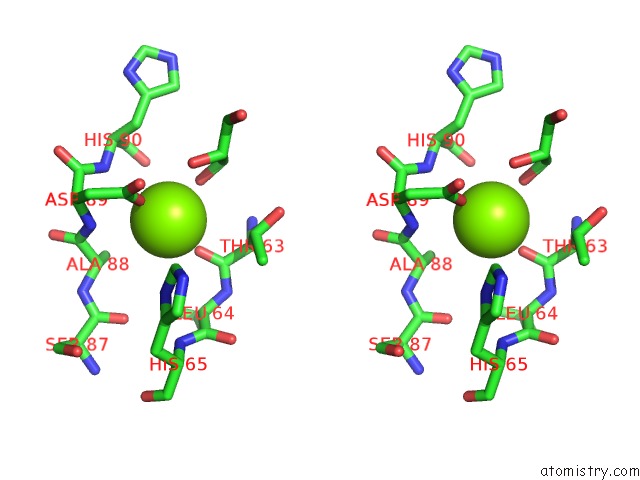
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Mouse MTH1 Mutant L116M with Inhibitor TH588 within 5.0Å range:
|
Reference:
M.Narwal,
A.S.Jemth,
R.Gustafsson,
I.Almlof,
U.Warpman Berglund,
T.Helleday,
P.Stenmark.
Crystal Structures and Inhibitor Interactions of Mouse and Dog MTH1 Reveal Species-Specific Differences in Affinity. Biochemistry V. 57 593 2018.
ISSN: ISSN 1520-4995
PubMed: 29281266
DOI: 10.1021/ACS.BIOCHEM.7B01163
Page generated: Mon Sep 30 23:57:39 2024
ISSN: ISSN 1520-4995
PubMed: 29281266
DOI: 10.1021/ACS.BIOCHEM.7B01163
Last articles
Cl in 5XQDCl in 5XPE
Cl in 5XNL
Cl in 5XP7
Cl in 5XNQ
Cl in 5XNV
Cl in 5XNA
Cl in 5XMV
Cl in 5XII
Cl in 5XN7