Magnesium »
PDB 6f6u-6fch »
6f95 »
Magnesium in PDB 6f95: Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A
(pdb code 6f95). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 5 binding sites of Magnesium where determined in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A, PDB code: 6f95:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Magnesium where determined in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A, PDB code: 6f95:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5;
Magnesium binding site 1 out of 5 in 6f95
Go back to
Magnesium binding site 1 out
of 5 in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A
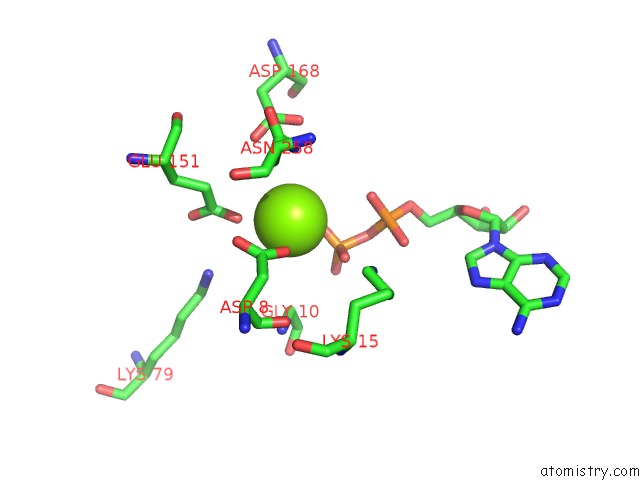
Mono view
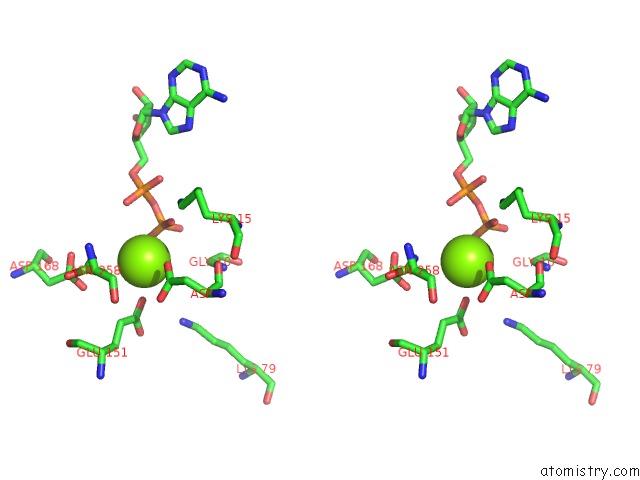
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A within 5.0Å range:
|
Magnesium binding site 2 out of 5 in 6f95
Go back to
Magnesium binding site 2 out
of 5 in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A
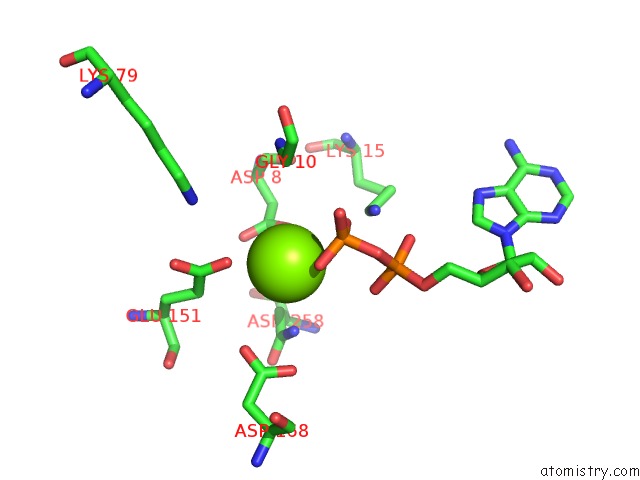
Mono view
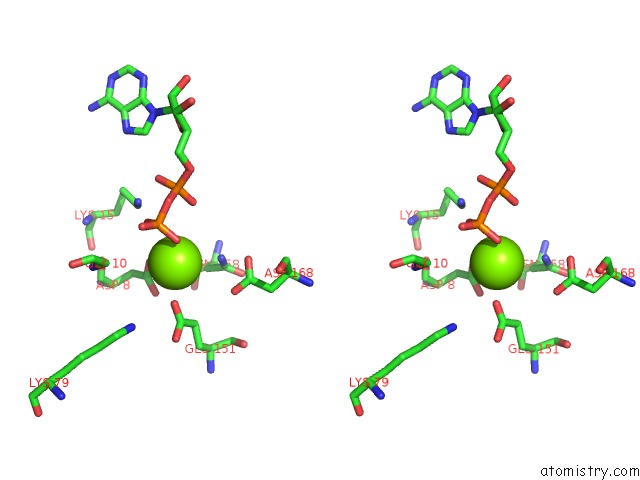
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A within 5.0Å range:
|
Magnesium binding site 3 out of 5 in 6f95
Go back to
Magnesium binding site 3 out
of 5 in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A
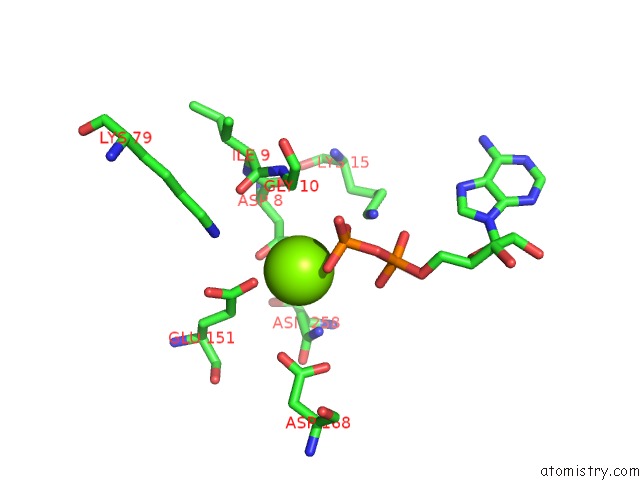
Mono view
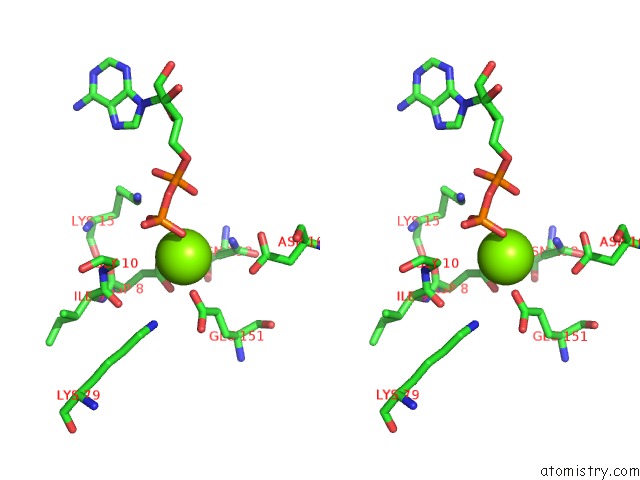
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A within 5.0Å range:
|
Magnesium binding site 4 out of 5 in 6f95
Go back to
Magnesium binding site 4 out
of 5 in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A
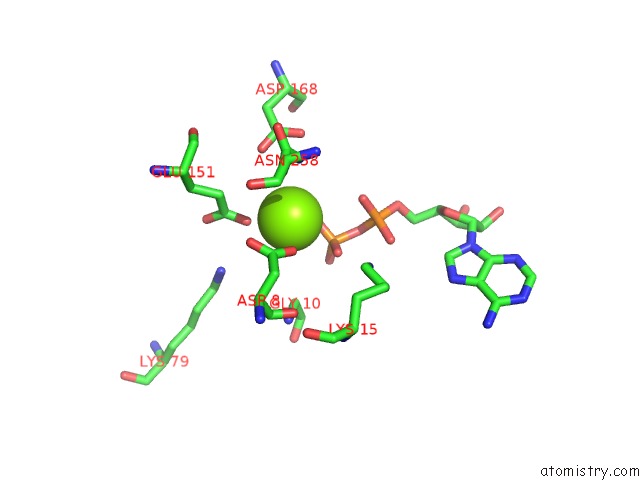
Mono view
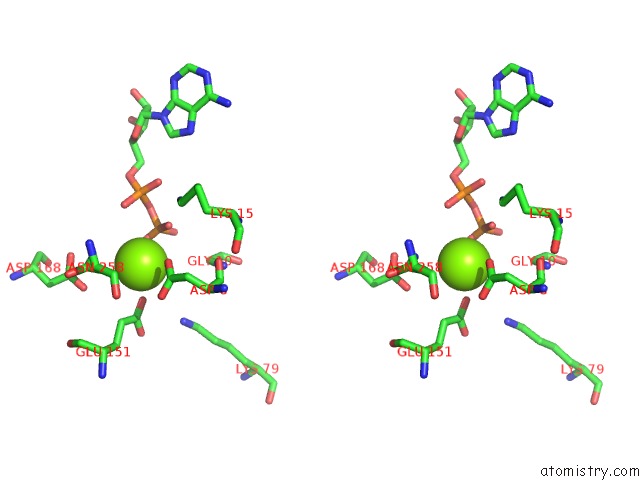
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A within 5.0Å range:
|
Magnesium binding site 5 out of 5 in 6f95
Go back to
Magnesium binding site 5 out
of 5 in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A
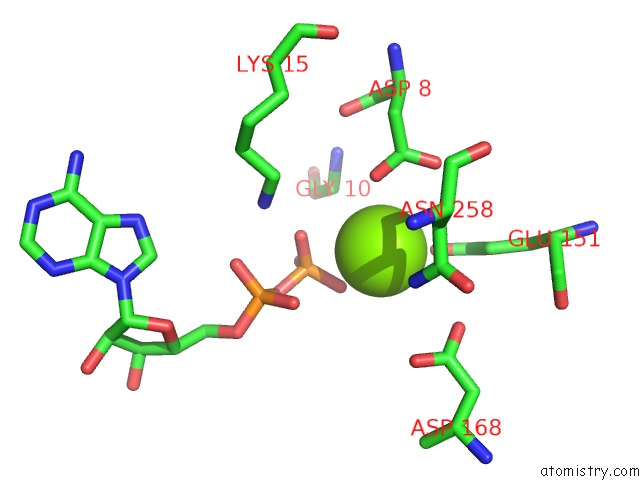
Mono view
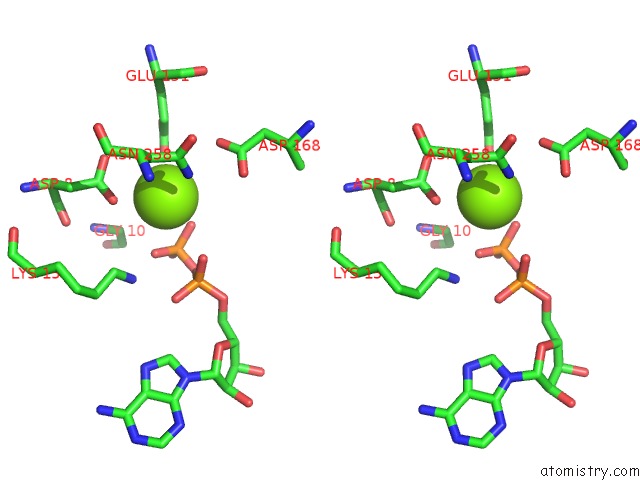
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A within 5.0Å range:
|
Reference:
A.Szewczak-Harris,
J.Lowe.
Cryo-Em Reconstruction of Alfa Frombacillus Subtilisreveals the Structure of A Simplified Actin-Like Filament at 3.4- Angstrom Resolution. Proc. Natl. Acad. Sci. V. 115 3458 2018U.S.A..
ISSN: ESSN 1091-6490
PubMed: 29440489
DOI: 10.1073/PNAS.1716424115
Page generated: Tue Oct 1 00:14:09 2024
ISSN: ESSN 1091-6490
PubMed: 29440489
DOI: 10.1073/PNAS.1716424115
Last articles
Cl in 5WYNCl in 5WW9
Cl in 5WW8
Cl in 5WW7
Cl in 5WW6
Cl in 5WW5
Cl in 5WW4
Cl in 5WW3
Cl in 5WU4
Cl in 5WU3