Magnesium »
PDB 6fkf-6ftv »
6flp »
Magnesium in PDB 6flp: Cryoem Structure of E.Coli Rna Polymerase Paused Elongation Complex Without Rna Hairpin Bound to Nusa
Enzymatic activity of Cryoem Structure of E.Coli Rna Polymerase Paused Elongation Complex Without Rna Hairpin Bound to Nusa
All present enzymatic activity of Cryoem Structure of E.Coli Rna Polymerase Paused Elongation Complex Without Rna Hairpin Bound to Nusa:
2.7.7.6;
2.7.7.6;
Other elements in 6flp:
The structure of Cryoem Structure of E.Coli Rna Polymerase Paused Elongation Complex Without Rna Hairpin Bound to Nusa also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryoem Structure of E.Coli Rna Polymerase Paused Elongation Complex Without Rna Hairpin Bound to Nusa
(pdb code 6flp). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Cryoem Structure of E.Coli Rna Polymerase Paused Elongation Complex Without Rna Hairpin Bound to Nusa, PDB code: 6flp:
In total only one binding site of Magnesium was determined in the Cryoem Structure of E.Coli Rna Polymerase Paused Elongation Complex Without Rna Hairpin Bound to Nusa, PDB code: 6flp:
Magnesium binding site 1 out of 1 in 6flp
Go back to
Magnesium binding site 1 out
of 1 in the Cryoem Structure of E.Coli Rna Polymerase Paused Elongation Complex Without Rna Hairpin Bound to Nusa
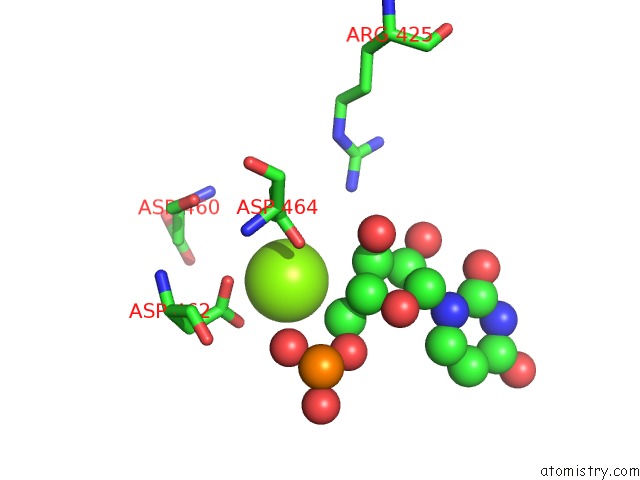
Mono view
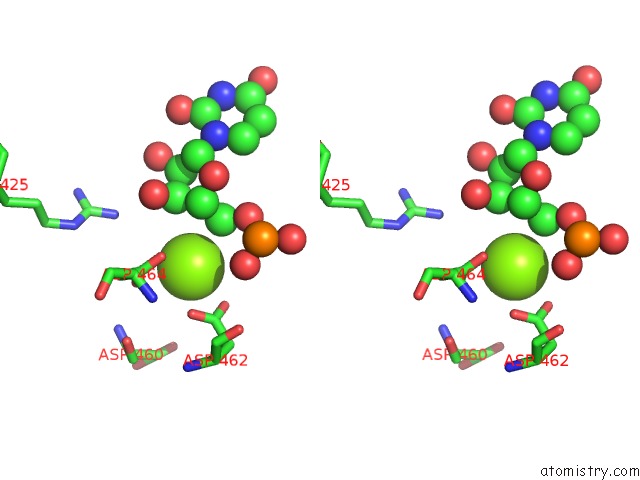
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryoem Structure of E.Coli Rna Polymerase Paused Elongation Complex Without Rna Hairpin Bound to Nusa within 5.0Å range:
|
Reference:
X.Guo,
A.G.Myasnikov,
J.Chen,
C.Crucifix,
G.Papai,
M.Takacs,
P.Schultz,
A.Weixlbaumer.
Structural Basis For Nusa Stabilized Transcriptional Pausing. Mol. Cell V. 69 816 2018.
ISSN: ISSN 1097-4164
PubMed: 29499136
DOI: 10.1016/J.MOLCEL.2018.02.008
Page generated: Wed Aug 13 06:09:12 2025
ISSN: ISSN 1097-4164
PubMed: 29499136
DOI: 10.1016/J.MOLCEL.2018.02.008
Last articles
Mg in 7ADSMg in 7ADE
Mg in 7ADI
Mg in 7ADD
Mg in 7ADC
Mg in 7ADB
Mg in 7AD9
Mg in 7ACQ
Mg in 7ACF
Mg in 7ACA