Magnesium »
PDB 6g7d-6gg6 »
6gej »
Magnesium in PDB 6gej: Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution.
Enzymatic activity of Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution.
All present enzymatic activity of Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution.:
3.6.4.12;
3.6.4.12;
Other elements in 6gej:
The structure of Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution. also contains other interesting chemical elements:
| Fluorine | (F) | 6 atoms |
| Zinc | (Zn) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution.
(pdb code 6gej). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 8 binding sites of Magnesium where determined in the Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution., PDB code: 6gej:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Magnesium where determined in the Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution., PDB code: 6gej:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
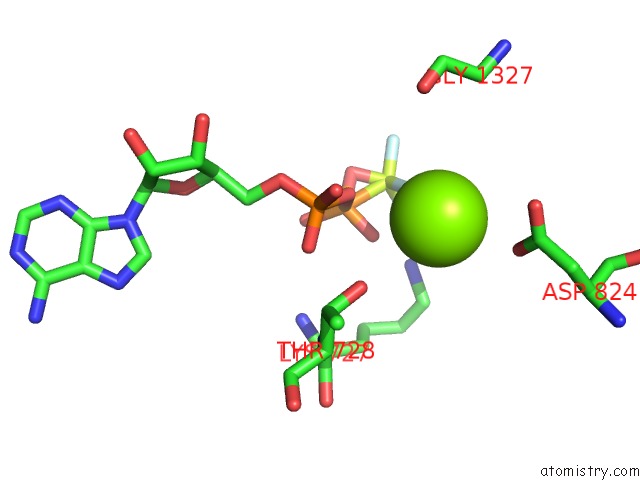
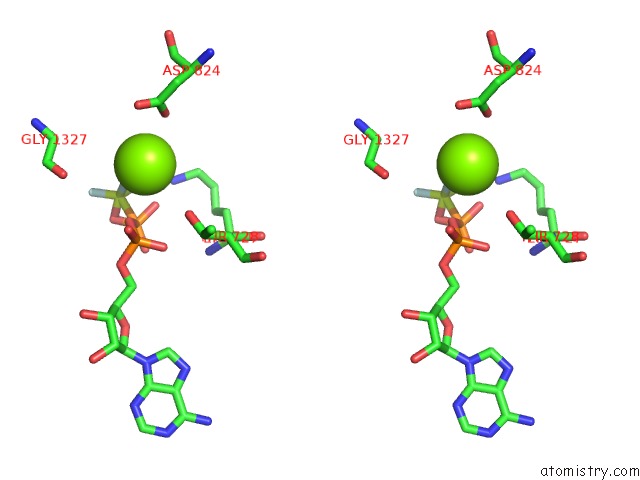
Magnesium binding site 1 out of 8 in 6gej
Go back to
Magnesium binding site 1 out
of 8 in the Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution.
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution. within 5.0Å range:
|
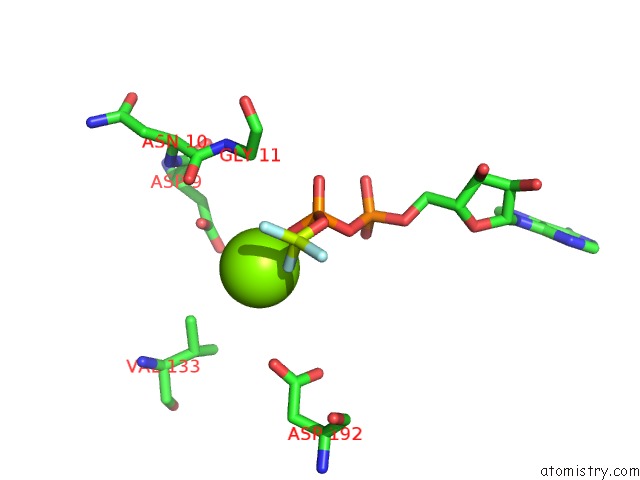
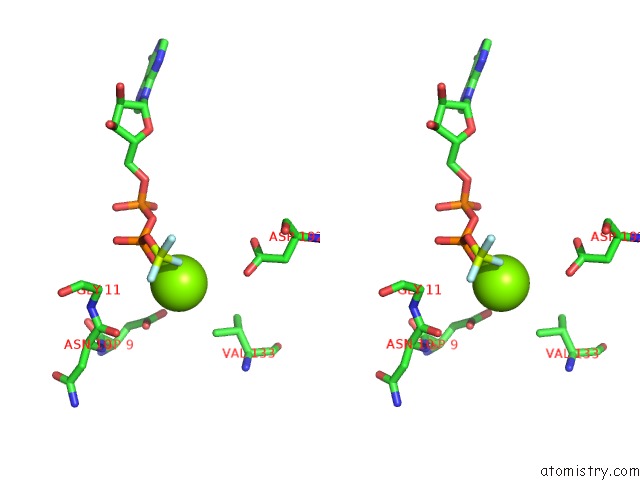
Magnesium binding site 2 out of 8 in 6gej
Go back to
Magnesium binding site 2 out
of 8 in the Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution.
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution. within 5.0Å range:
|
Magnesium binding site 3 out of 8 in 6gej
Go back to
Magnesium binding site 3 out
of 8 in the Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution.
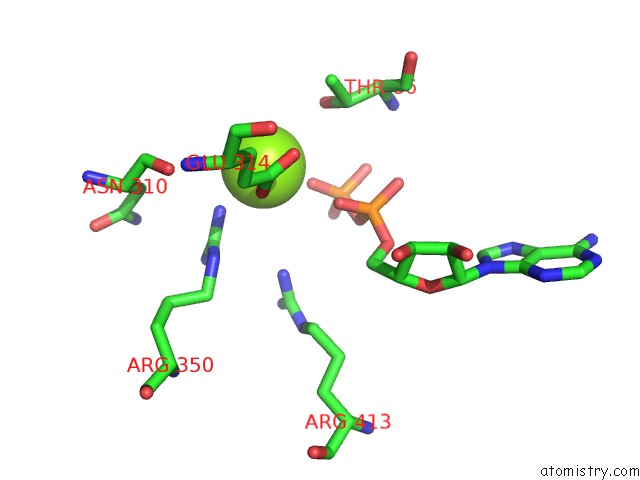
Mono view
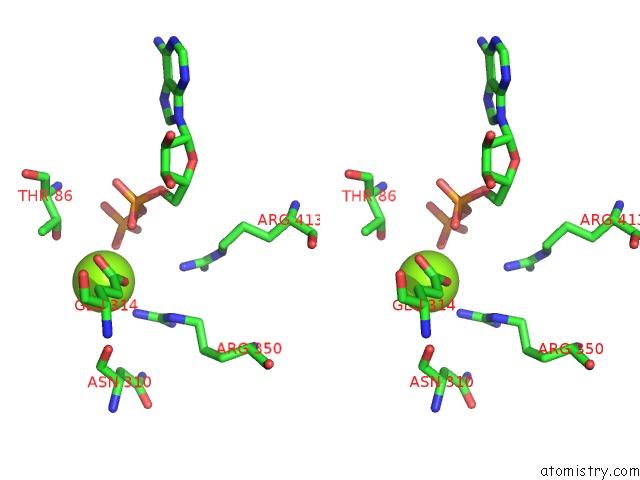
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution. within 5.0Å range:
|
Magnesium binding site 4 out of 8 in 6gej
Go back to
Magnesium binding site 4 out
of 8 in the Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution.
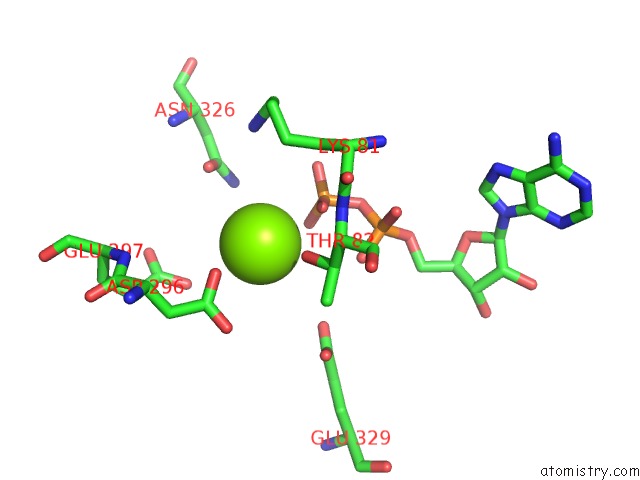
Mono view
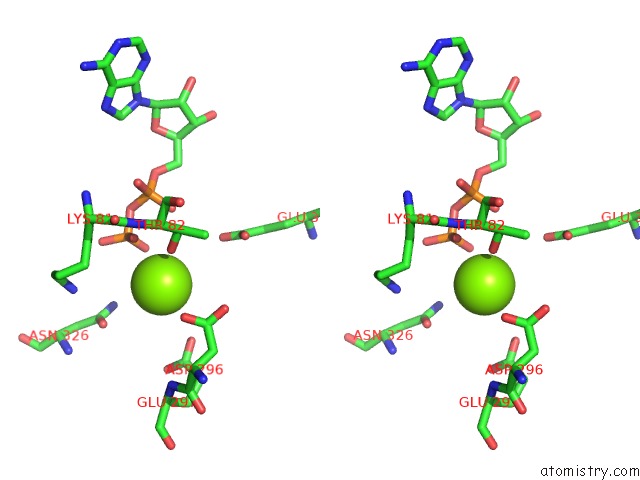
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution. within 5.0Å range:
|
Magnesium binding site 5 out of 8 in 6gej
Go back to
Magnesium binding site 5 out
of 8 in the Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution.
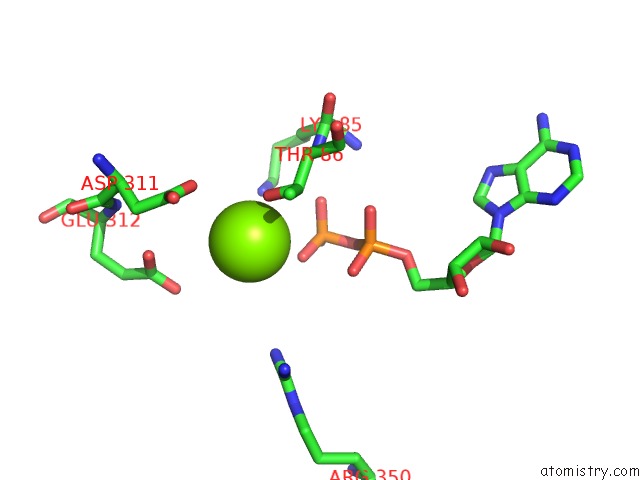
Mono view
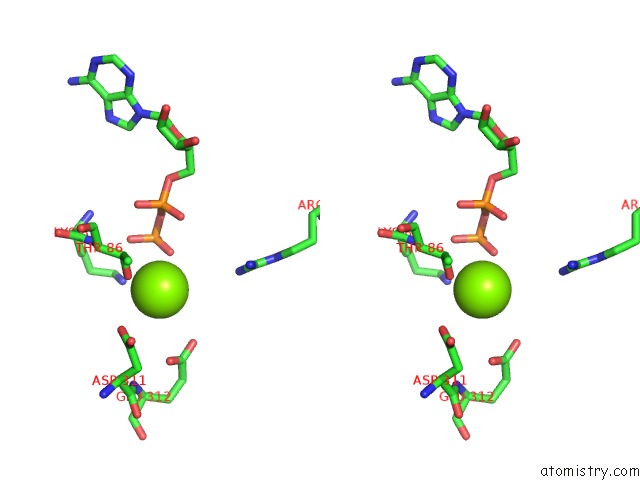
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution. within 5.0Å range:
|
Magnesium binding site 6 out of 8 in 6gej
Go back to
Magnesium binding site 6 out
of 8 in the Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution.
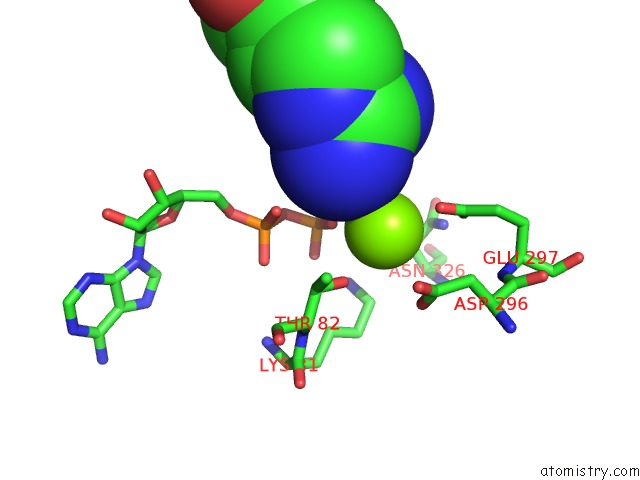
Mono view
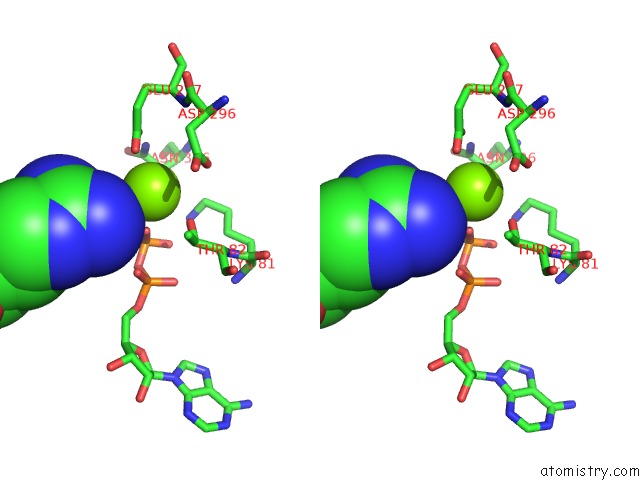
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 6 of Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution. within 5.0Å range:
|
Magnesium binding site 7 out of 8 in 6gej
Go back to
Magnesium binding site 7 out
of 8 in the Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution.
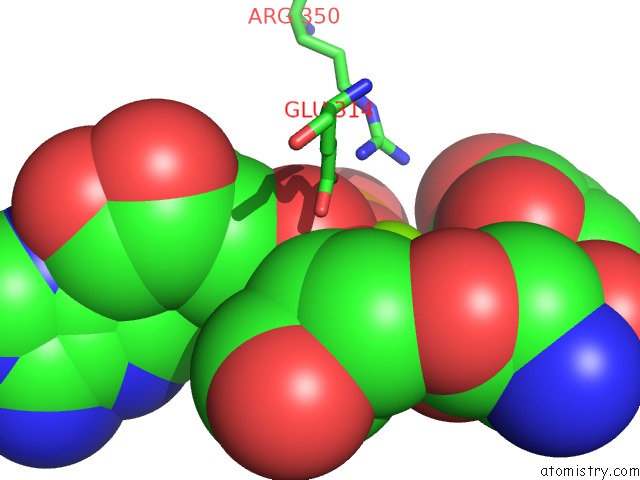
Mono view
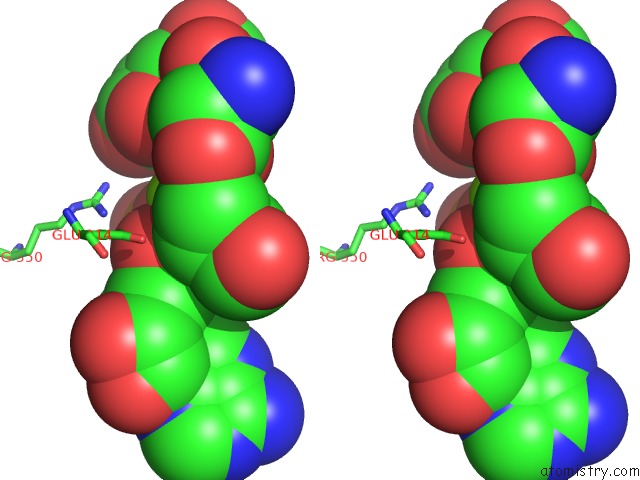
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 7 of Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution. within 5.0Å range:
|
Magnesium binding site 8 out of 8 in 6gej
Go back to
Magnesium binding site 8 out
of 8 in the Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution.
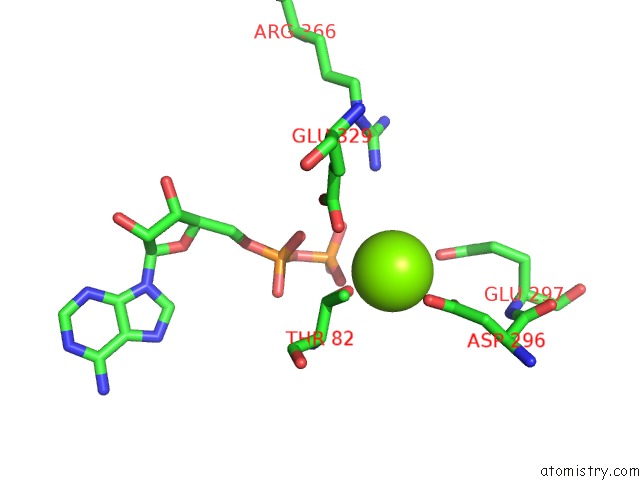
Mono view
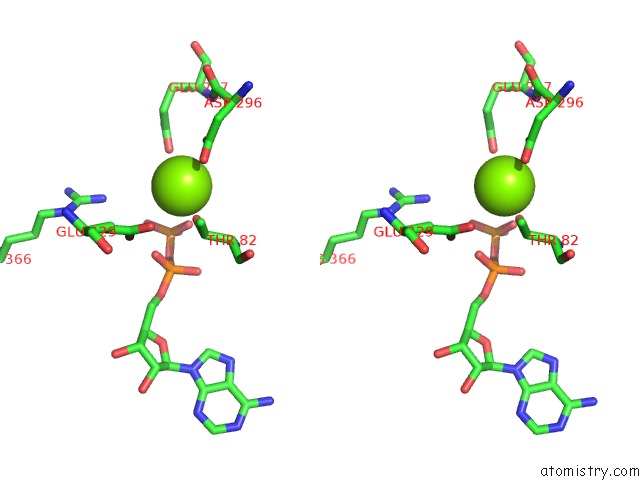
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 8 of Chromatin Remodeller-Nucleosome Complex at 3.6 A Resolution. within 5.0Å range:
|
Reference:
O.Willhoft,
M.Ghoneim,
C.L.Lin,
E.Y.D.Chua,
M.Wilkinson,
Y.Chaban,
R.Ayala,
E.A.Mccormack,
L.Ocloo,
D.S.Rueda,
D.B.Wigley.
Structure and Dynamics of the Yeast SWR1-Nucleosome Complex. Science V. 362 2018.
ISSN: ESSN 1095-9203
PubMed: 30309918
DOI: 10.1126/SCIENCE.AAT7716
Page generated: Tue Oct 1 01:07:03 2024
ISSN: ESSN 1095-9203
PubMed: 30309918
DOI: 10.1126/SCIENCE.AAT7716
Last articles
F in 4JAVF in 4JA8
F in 4JBY
F in 4JB4
F in 4JAS
F in 4JA2
F in 4J3J
F in 4J8W
F in 4J8M
F in 4J6A