Magnesium »
PDB 6h5e-6hdi »
6hde »
Magnesium in PDB 6hde: Structure of Escherichia Coli Dutpase Q93H Mutant
Enzymatic activity of Structure of Escherichia Coli Dutpase Q93H Mutant
All present enzymatic activity of Structure of Escherichia Coli Dutpase Q93H Mutant:
3.6.1.23;
3.6.1.23;
Protein crystallography data
The structure of Structure of Escherichia Coli Dutpase Q93H Mutant, PDB code: 6hde
was solved by
A.Benedek,
B.G.Vertessy,
I.Leveles,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.81 / 1.82 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 63.200, 66.500, 95.300, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.1 / 22.1 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of Escherichia Coli Dutpase Q93H Mutant
(pdb code 6hde). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Structure of Escherichia Coli Dutpase Q93H Mutant, PDB code: 6hde:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Structure of Escherichia Coli Dutpase Q93H Mutant, PDB code: 6hde:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 6hde
Go back to
Magnesium binding site 1 out
of 3 in the Structure of Escherichia Coli Dutpase Q93H Mutant
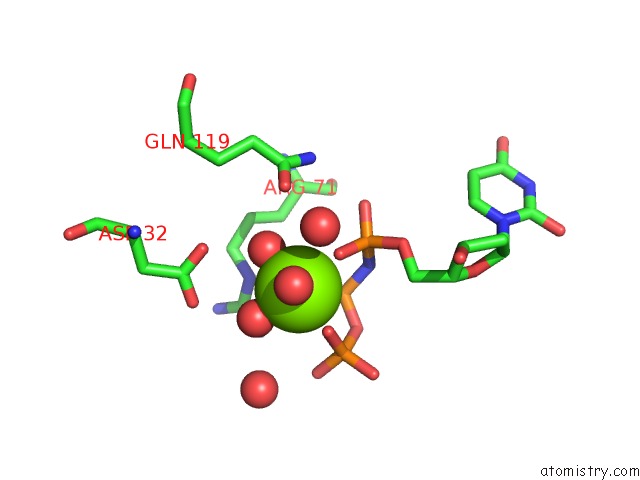
Mono view
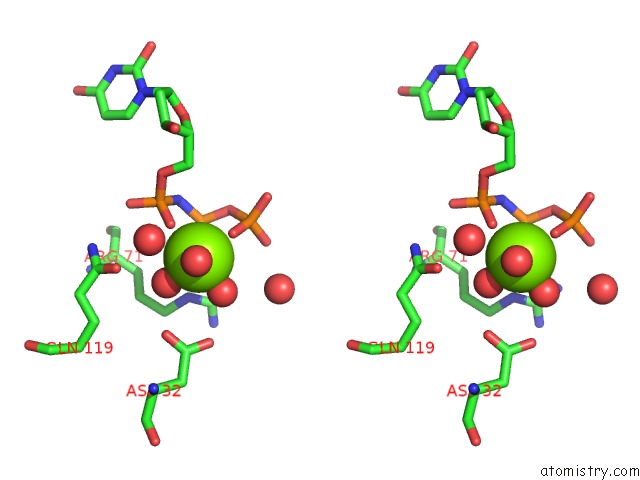
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of Escherichia Coli Dutpase Q93H Mutant within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 6hde
Go back to
Magnesium binding site 2 out
of 3 in the Structure of Escherichia Coli Dutpase Q93H Mutant
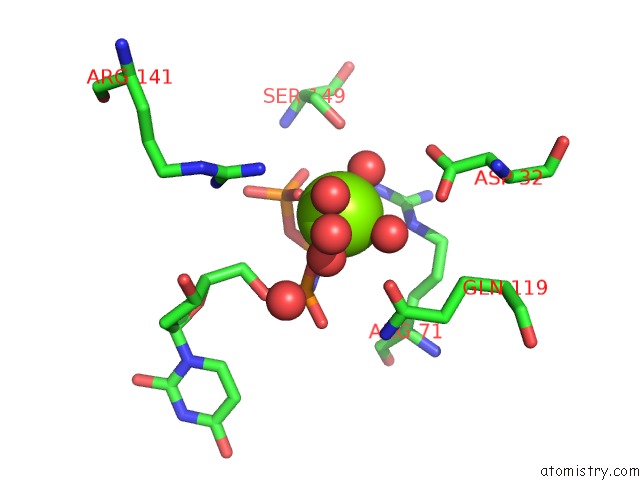
Mono view
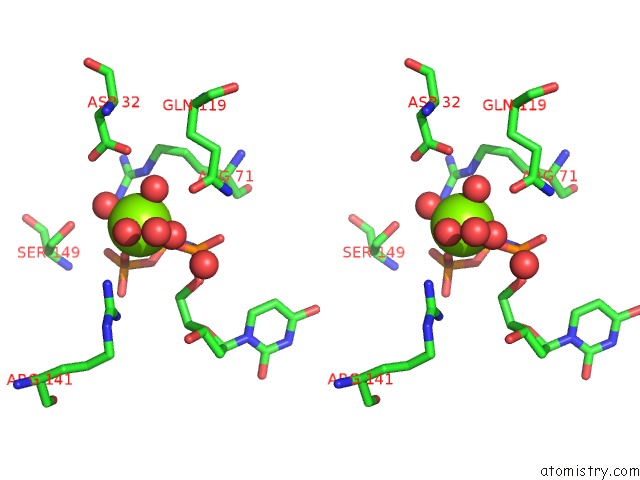
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of Escherichia Coli Dutpase Q93H Mutant within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 6hde
Go back to
Magnesium binding site 3 out
of 3 in the Structure of Escherichia Coli Dutpase Q93H Mutant
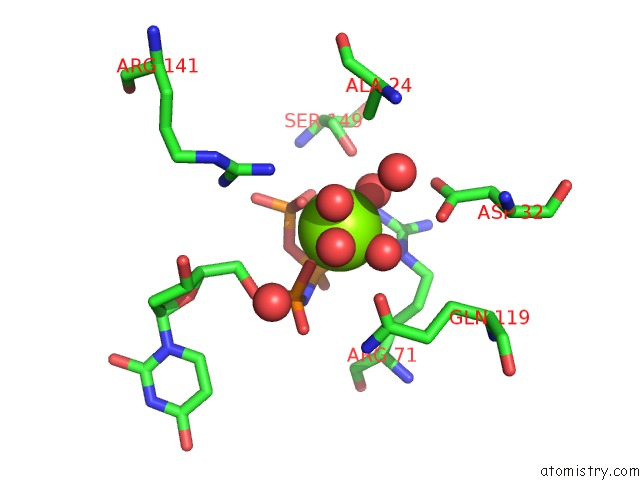
Mono view
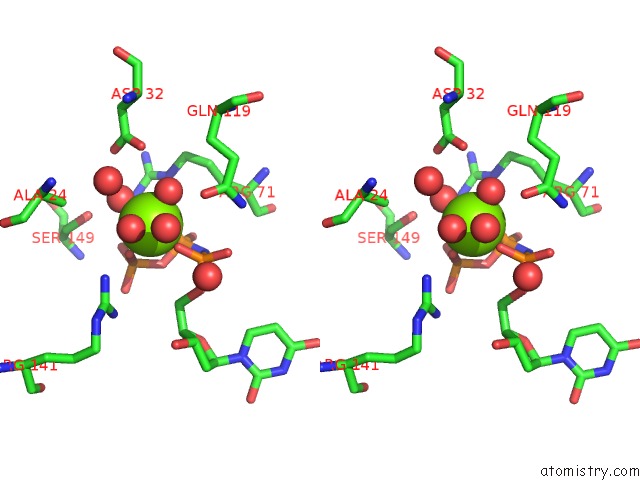
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structure of Escherichia Coli Dutpase Q93H Mutant within 5.0Å range:
|
Reference:
A.Benedek,
F.Temesvary-Kis,
T.Khatanbaatar,
I.Leveles,
E.V.Suranyi,
J.E.Szabo,
L.Wunderlich,
B.G.Vertessy.
The Role of A Key Amino Acid Position in Species-Specific Proteinaceous Dutpase Inhibition. Biomolecules V. 9 2019.
ISSN: ESSN 2218-273X
PubMed: 31174420
DOI: 10.3390/BIOM9060221
Page generated: Wed Aug 13 06:57:02 2025
ISSN: ESSN 2218-273X
PubMed: 31174420
DOI: 10.3390/BIOM9060221
Last articles
Mg in 7EG8Mg in 7EG7
Mg in 7EG4
Mg in 7EG1
Mg in 7EG0
Mg in 7EFN
Mg in 7EFM
Mg in 7EFL
Mg in 7EDZ
Mg in 7EF6