Magnesium »
PDB 6hvy-6i1k »
6hz4 »
Magnesium in PDB 6hz4: Structure of Mcrbc Without Dna Binding Domains (One Half of the Full Complex)
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of Mcrbc Without Dna Binding Domains (One Half of the Full Complex)
(pdb code 6hz4). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Structure of Mcrbc Without Dna Binding Domains (One Half of the Full Complex), PDB code: 6hz4:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Structure of Mcrbc Without Dna Binding Domains (One Half of the Full Complex), PDB code: 6hz4:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 6hz4
Go back to
Magnesium binding site 1 out
of 3 in the Structure of Mcrbc Without Dna Binding Domains (One Half of the Full Complex)
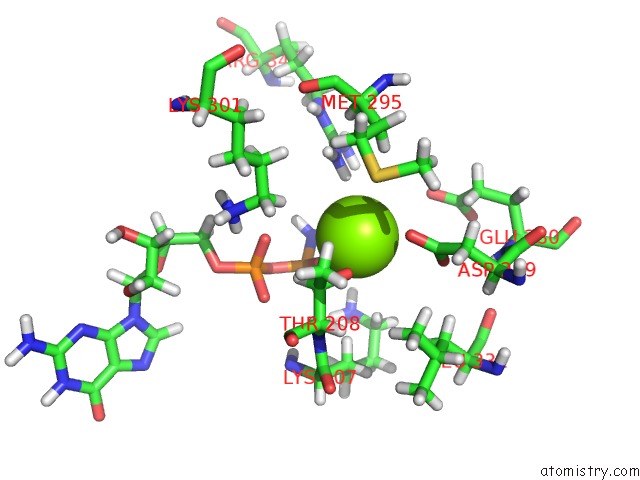
Mono view
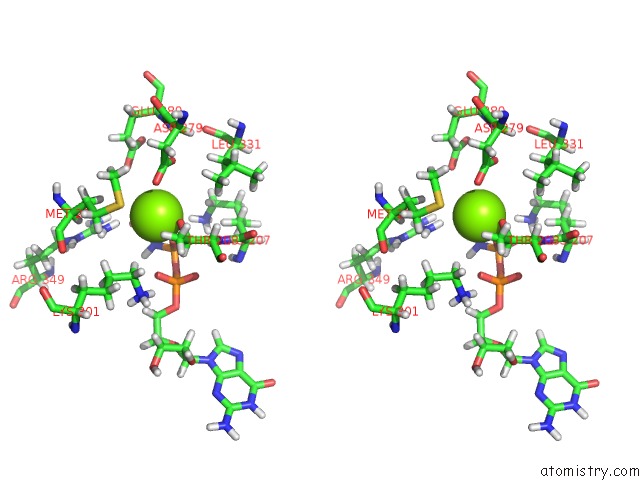
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of Mcrbc Without Dna Binding Domains (One Half of the Full Complex) within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 6hz4
Go back to
Magnesium binding site 2 out
of 3 in the Structure of Mcrbc Without Dna Binding Domains (One Half of the Full Complex)
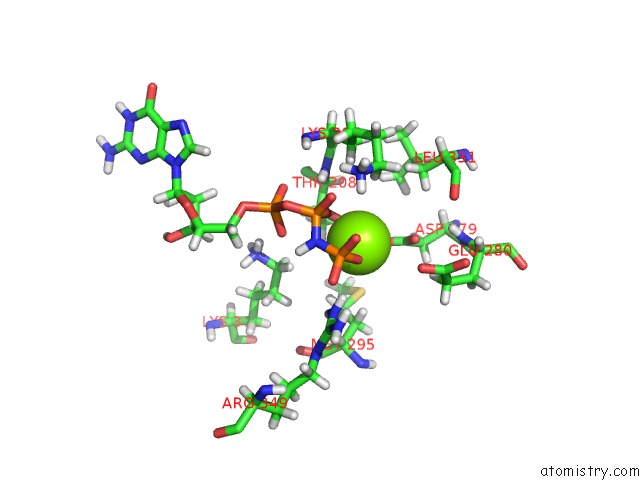
Mono view
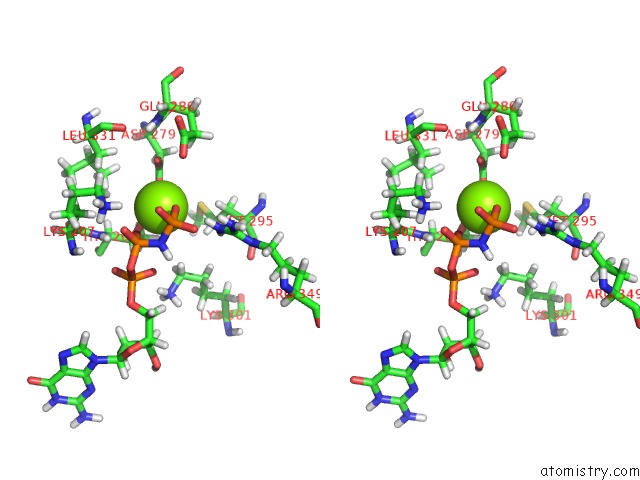
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of Mcrbc Without Dna Binding Domains (One Half of the Full Complex) within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 6hz4
Go back to
Magnesium binding site 3 out
of 3 in the Structure of Mcrbc Without Dna Binding Domains (One Half of the Full Complex)
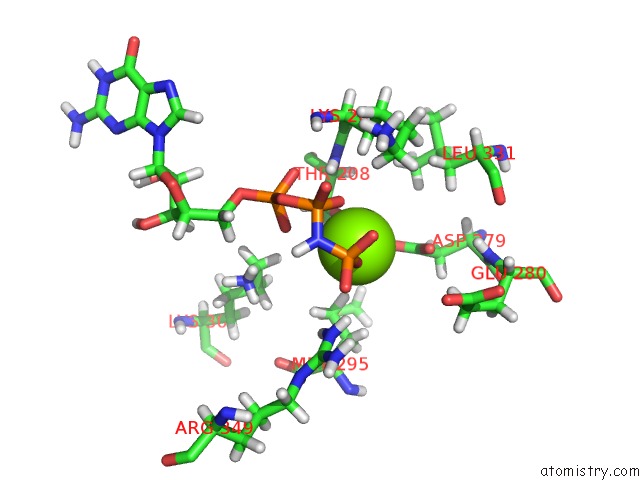
Mono view
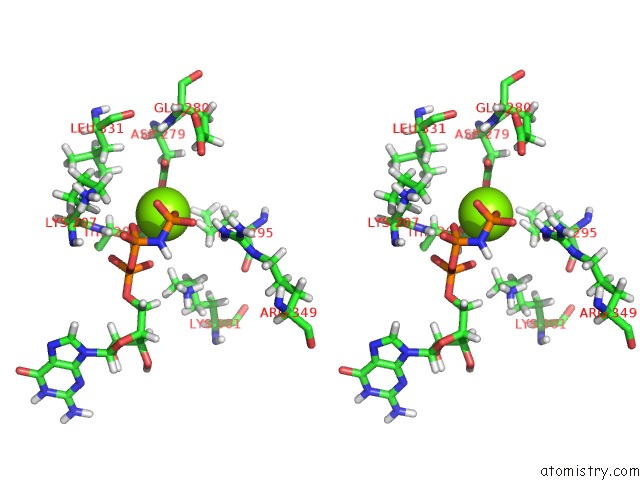
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structure of Mcrbc Without Dna Binding Domains (One Half of the Full Complex) within 5.0Å range:
|
Reference:
N.Nirwan,
Y.Itoh,
P.Singh,
S.Bandyopadhyay,
K.R.Vinothkumar,
A.Amunts,
K.Saikrishnan.
Structure-Based Mechanism For Activation of the Aaa+ Gtpase Mcrb By the Endonuclease Mcrc. Nat Commun V. 10 3058 2019.
ISSN: ESSN 2041-1723
PubMed: 31296862
DOI: 10.1038/S41467-019-11084-1
Page generated: Wed Aug 13 07:44:37 2025
ISSN: ESSN 2041-1723
PubMed: 31296862
DOI: 10.1038/S41467-019-11084-1
Last articles
Mg in 7EQ2Mg in 7EOP
Mg in 7EPU
Mg in 7EOO
Mg in 7EON
Mg in 7EPK
Mg in 7EOL
Mg in 7EOK
Mg in 7EOJ
Mg in 7EOM