Magnesium »
PDB 6iag-6ik9 »
6ihu »
Magnesium in PDB 6ihu: Crystal Structure of Bacterial Serine Phosphatase Bearing R161A Mutation
Enzymatic activity of Crystal Structure of Bacterial Serine Phosphatase Bearing R161A Mutation
All present enzymatic activity of Crystal Structure of Bacterial Serine Phosphatase Bearing R161A Mutation:
3.1.3.16;
3.1.3.16;
Protein crystallography data
The structure of Crystal Structure of Bacterial Serine Phosphatase Bearing R161A Mutation, PDB code: 6ihu
was solved by
C.-G.Yang,
T.Yang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.48 / 1.84 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 46.998, 37.745, 65.575, 90.00, 102.87, 90.00 |
| R / Rfree (%) | 16.9 / 20 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Bacterial Serine Phosphatase Bearing R161A Mutation
(pdb code 6ihu). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Crystal Structure of Bacterial Serine Phosphatase Bearing R161A Mutation, PDB code: 6ihu:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Crystal Structure of Bacterial Serine Phosphatase Bearing R161A Mutation, PDB code: 6ihu:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 6ihu
Go back to
Magnesium binding site 1 out
of 3 in the Crystal Structure of Bacterial Serine Phosphatase Bearing R161A Mutation
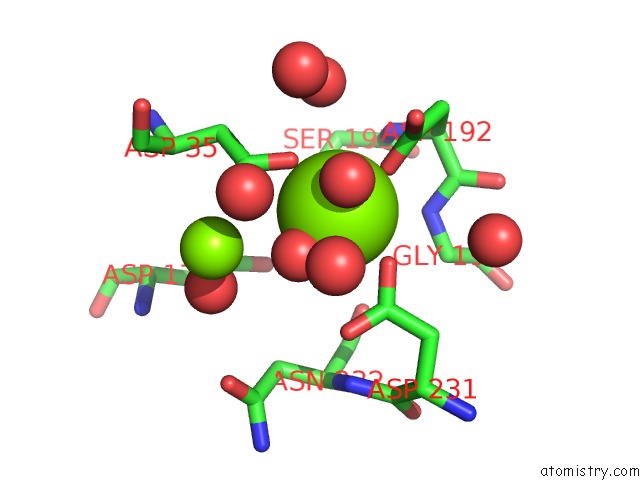
Mono view
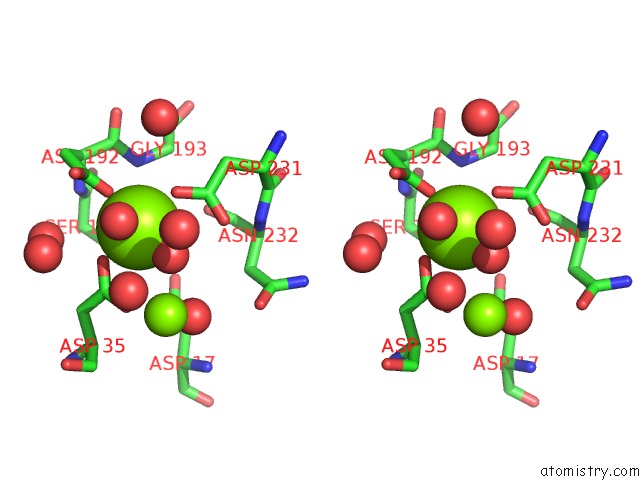
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Bacterial Serine Phosphatase Bearing R161A Mutation within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 6ihu
Go back to
Magnesium binding site 2 out
of 3 in the Crystal Structure of Bacterial Serine Phosphatase Bearing R161A Mutation
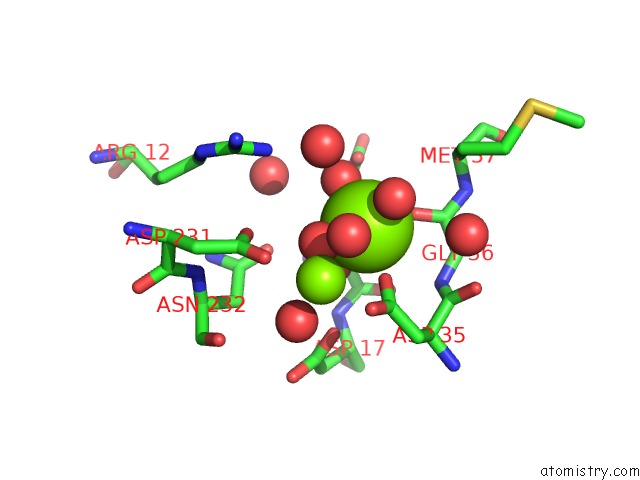
Mono view
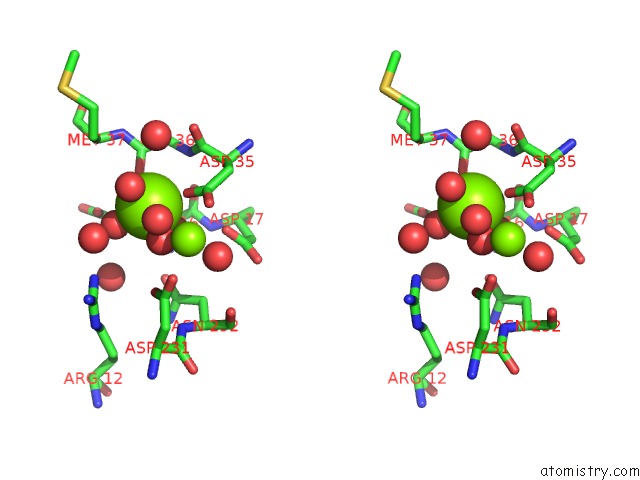
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Bacterial Serine Phosphatase Bearing R161A Mutation within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 6ihu
Go back to
Magnesium binding site 3 out
of 3 in the Crystal Structure of Bacterial Serine Phosphatase Bearing R161A Mutation
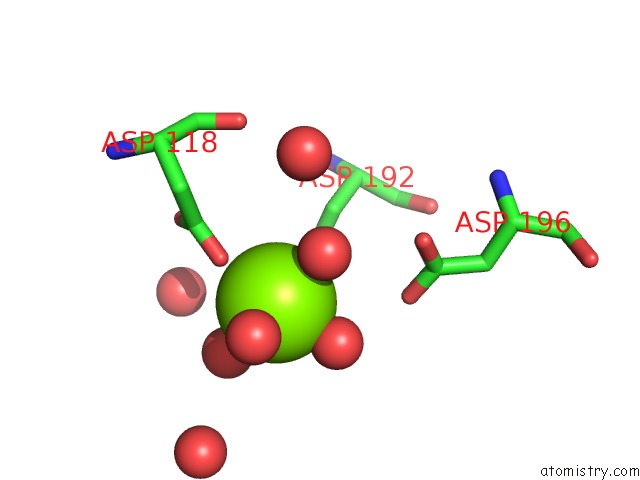
Mono view
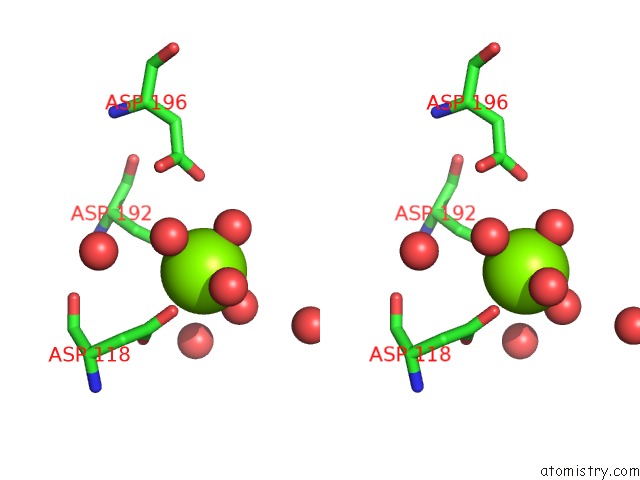
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of Bacterial Serine Phosphatase Bearing R161A Mutation within 5.0Å range:
|
Reference:
T.Yang,
T.Liu,
J.Gan,
K.Yu,
K.Chen,
W.Xue,
L.Lan,
S.Yang,
C.G.Yang.
Structural Insight Into the Mechanism of Staphylococcus Aureus STP1 Phosphatase. Acs Infect Dis. V. 5 841 2019.
ISSN: ESSN 2373-8227
PubMed: 30868877
DOI: 10.1021/ACSINFECDIS.8B00316
Page generated: Wed Aug 13 08:08:07 2025
ISSN: ESSN 2373-8227
PubMed: 30868877
DOI: 10.1021/ACSINFECDIS.8B00316
Last articles
Mg in 7EXCMg in 7EWB
Mg in 7EXP
Mg in 7EW6
Mg in 7EQD
Mg in 7EWA
Mg in 7EW9
Mg in 7EV7
Mg in 7EVI
Mg in 7EUQ