Magnesium »
PDB 6m69-6mlw »
6mfw »
Magnesium in PDB 6mfw: Crystal Structure of A 4-Domain Construct of Lgra in the Substrate Donation State
Protein crystallography data
The structure of Crystal Structure of A 4-Domain Construct of Lgra in the Substrate Donation State, PDB code: 6mfw
was solved by
J.M.Reimer,
M.Eivaskhani,
T.M.Schmeing,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 69.33 / 2.50 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 66.370, 133.870, 162.099, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20 / 24.4 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of A 4-Domain Construct of Lgra in the Substrate Donation State
(pdb code 6mfw). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of A 4-Domain Construct of Lgra in the Substrate Donation State, PDB code: 6mfw:
In total only one binding site of Magnesium was determined in the Crystal Structure of A 4-Domain Construct of Lgra in the Substrate Donation State, PDB code: 6mfw:
Magnesium binding site 1 out of 1 in 6mfw
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of A 4-Domain Construct of Lgra in the Substrate Donation State
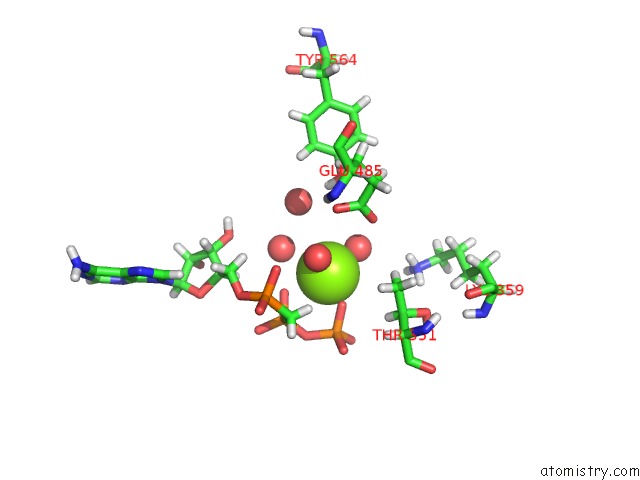
Mono view
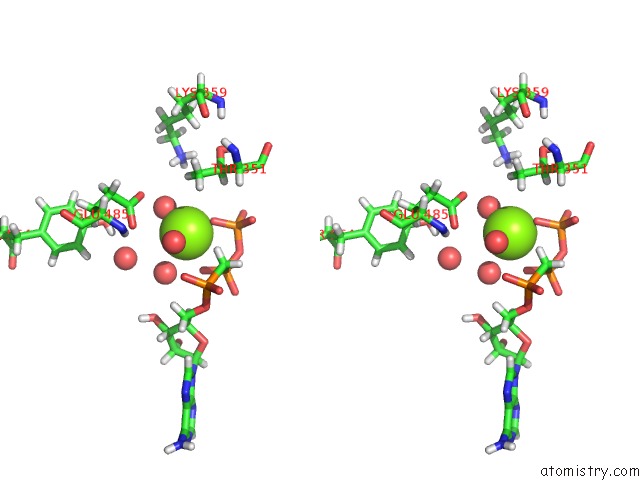
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of A 4-Domain Construct of Lgra in the Substrate Donation State within 5.0Å range:
|
Reference:
J.M.Reimer,
M.Eivaskhani,
I.Harb,
A.Guarne,
M.Weigt,
T.M.Schmeing.
Structures of A Dimodular Nonribosomal Peptide Synthetase Reveal Conformational Flexibility. Science V. 366 2019.
ISSN: ESSN 1095-9203
PubMed: 31699907
DOI: 10.1126/SCIENCE.AAW4388
Page generated: Tue Oct 1 11:37:13 2024
ISSN: ESSN 1095-9203
PubMed: 31699907
DOI: 10.1126/SCIENCE.AAW4388
Last articles
Ca in 5VT8Ca in 5VYF
Ca in 5VYB
Ca in 5VXZ
Ca in 5VTM
Ca in 5VWM
Ca in 5VTD
Ca in 5VUG
Ca in 5VS1
Ca in 5VRB