Magnesium »
PDB 6n7n-6nk8 »
6n7s »
Magnesium in PDB 6n7s: Structure of Bacteriophage T7 E343Q Mutant GP4 Helicase-Primase in Complex with Ssdna, Dttp, Ac Dinucleotide and Ctp (Form II)
Enzymatic activity of Structure of Bacteriophage T7 E343Q Mutant GP4 Helicase-Primase in Complex with Ssdna, Dttp, Ac Dinucleotide and Ctp (Form II)
All present enzymatic activity of Structure of Bacteriophage T7 E343Q Mutant GP4 Helicase-Primase in Complex with Ssdna, Dttp, Ac Dinucleotide and Ctp (Form II):
3.6.4.12;
3.6.4.12;
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of Bacteriophage T7 E343Q Mutant GP4 Helicase-Primase in Complex with Ssdna, Dttp, Ac Dinucleotide and Ctp (Form II)
(pdb code 6n7s). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Structure of Bacteriophage T7 E343Q Mutant GP4 Helicase-Primase in Complex with Ssdna, Dttp, Ac Dinucleotide and Ctp (Form II), PDB code: 6n7s:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Structure of Bacteriophage T7 E343Q Mutant GP4 Helicase-Primase in Complex with Ssdna, Dttp, Ac Dinucleotide and Ctp (Form II), PDB code: 6n7s:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 6n7s
Go back to
Magnesium binding site 1 out
of 4 in the Structure of Bacteriophage T7 E343Q Mutant GP4 Helicase-Primase in Complex with Ssdna, Dttp, Ac Dinucleotide and Ctp (Form II)
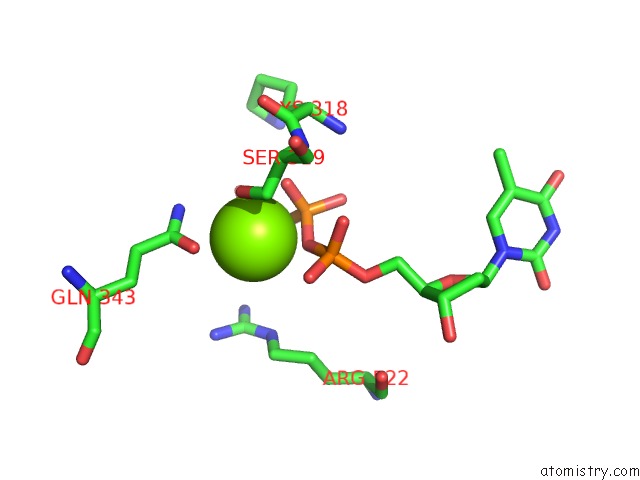
Mono view
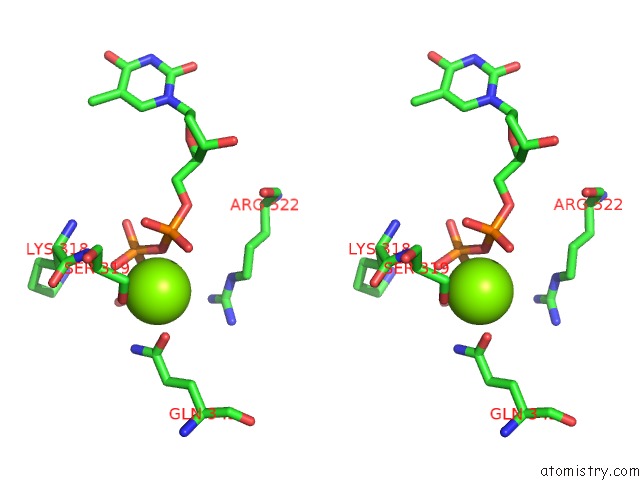
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of Bacteriophage T7 E343Q Mutant GP4 Helicase-Primase in Complex with Ssdna, Dttp, Ac Dinucleotide and Ctp (Form II) within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 6n7s
Go back to
Magnesium binding site 2 out
of 4 in the Structure of Bacteriophage T7 E343Q Mutant GP4 Helicase-Primase in Complex with Ssdna, Dttp, Ac Dinucleotide and Ctp (Form II)
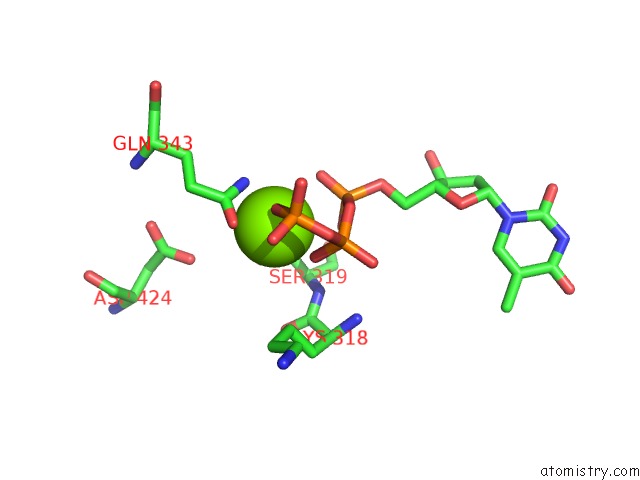
Mono view
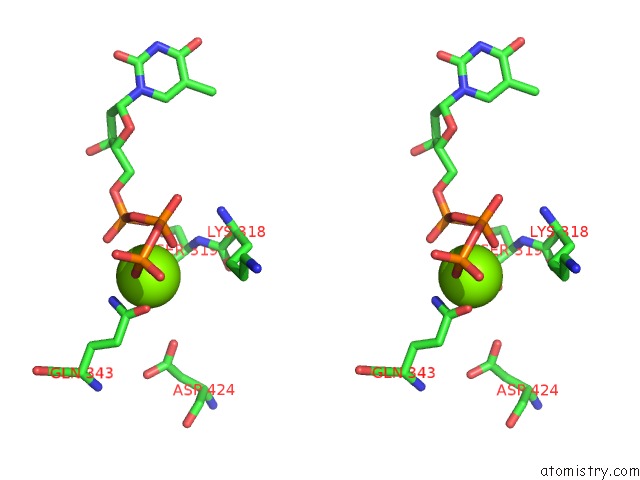
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of Bacteriophage T7 E343Q Mutant GP4 Helicase-Primase in Complex with Ssdna, Dttp, Ac Dinucleotide and Ctp (Form II) within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 6n7s
Go back to
Magnesium binding site 3 out
of 4 in the Structure of Bacteriophage T7 E343Q Mutant GP4 Helicase-Primase in Complex with Ssdna, Dttp, Ac Dinucleotide and Ctp (Form II)
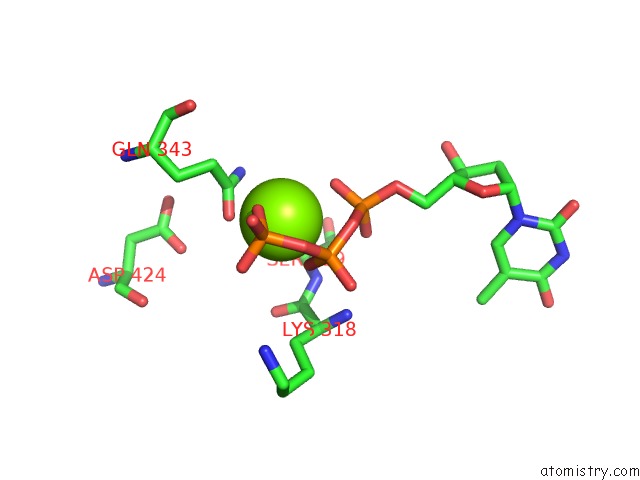
Mono view
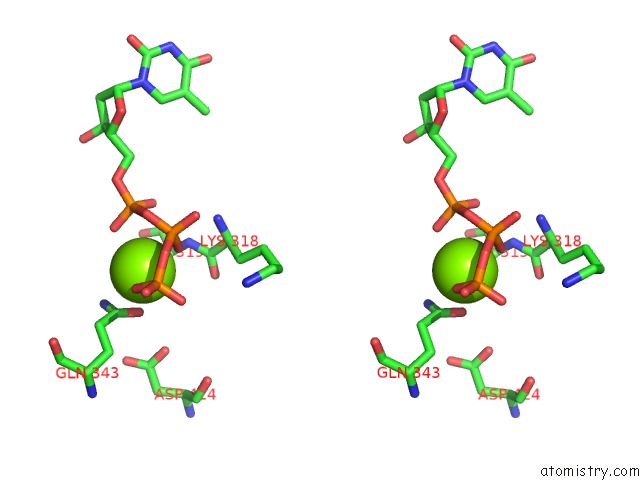
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structure of Bacteriophage T7 E343Q Mutant GP4 Helicase-Primase in Complex with Ssdna, Dttp, Ac Dinucleotide and Ctp (Form II) within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 6n7s
Go back to
Magnesium binding site 4 out
of 4 in the Structure of Bacteriophage T7 E343Q Mutant GP4 Helicase-Primase in Complex with Ssdna, Dttp, Ac Dinucleotide and Ctp (Form II)
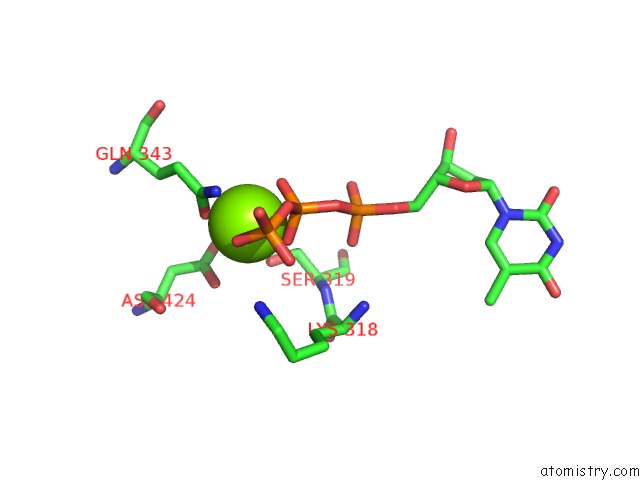
Mono view
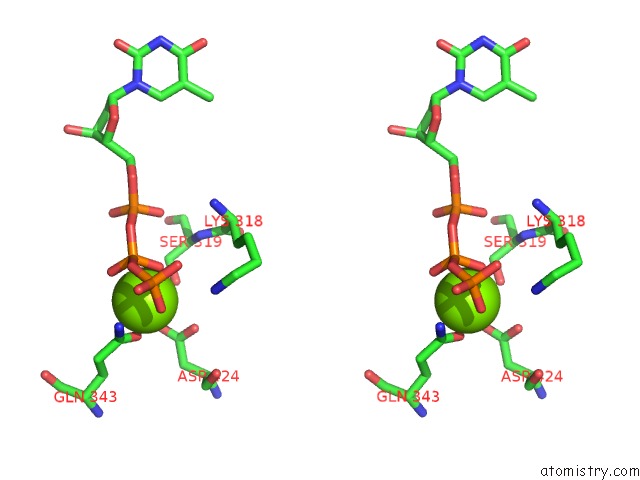
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Structure of Bacteriophage T7 E343Q Mutant GP4 Helicase-Primase in Complex with Ssdna, Dttp, Ac Dinucleotide and Ctp (Form II) within 5.0Å range:
|
Reference:
Y.Gao,
Y.Cui,
T.Fox,
S.Lin,
H.Wang,
N.De Val,
Z.H.Zhou,
W.Yang.
Structures and Operating Principles of the Replisome. Science V. 363 2019.
ISSN: ESSN 1095-9203
PubMed: 30679383
DOI: 10.1126/SCIENCE.AAV7003
Page generated: Tue Oct 1 12:34:29 2024
ISSN: ESSN 1095-9203
PubMed: 30679383
DOI: 10.1126/SCIENCE.AAV7003
Last articles
F in 4MC2F in 4MC1
F in 4M8N
F in 4MBS
F in 4MBJ
F in 4M6Q
F in 4M81
F in 4M7I
F in 4M5U
F in 4M3G