Magnesium »
PDB 6non-6o2p »
6ntd »
Magnesium in PDB 6ntd: Crystal Structure of G12V Hras-Gppnhp Bound in Complex with the Engineered Rbd Variant 12 of Craf Kinase Protein
Enzymatic activity of Crystal Structure of G12V Hras-Gppnhp Bound in Complex with the Engineered Rbd Variant 12 of Craf Kinase Protein
All present enzymatic activity of Crystal Structure of G12V Hras-Gppnhp Bound in Complex with the Engineered Rbd Variant 12 of Craf Kinase Protein:
2.7.11.1;
2.7.11.1;
Protein crystallography data
The structure of Crystal Structure of G12V Hras-Gppnhp Bound in Complex with the Engineered Rbd Variant 12 of Craf Kinase Protein, PDB code: 6ntd
was solved by
P.Maisonneuve,
I.Kurinov,
S.Wiechmann,
A.Ernst,
F.Sicheri,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 79.47 / 3.15 |
| Space group | P 63 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 91.761, 91.761, 151.590, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 27.3 / 28.7 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of G12V Hras-Gppnhp Bound in Complex with the Engineered Rbd Variant 12 of Craf Kinase Protein
(pdb code 6ntd). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of G12V Hras-Gppnhp Bound in Complex with the Engineered Rbd Variant 12 of Craf Kinase Protein, PDB code: 6ntd:
In total only one binding site of Magnesium was determined in the Crystal Structure of G12V Hras-Gppnhp Bound in Complex with the Engineered Rbd Variant 12 of Craf Kinase Protein, PDB code: 6ntd:
Magnesium binding site 1 out of 1 in 6ntd
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of G12V Hras-Gppnhp Bound in Complex with the Engineered Rbd Variant 12 of Craf Kinase Protein
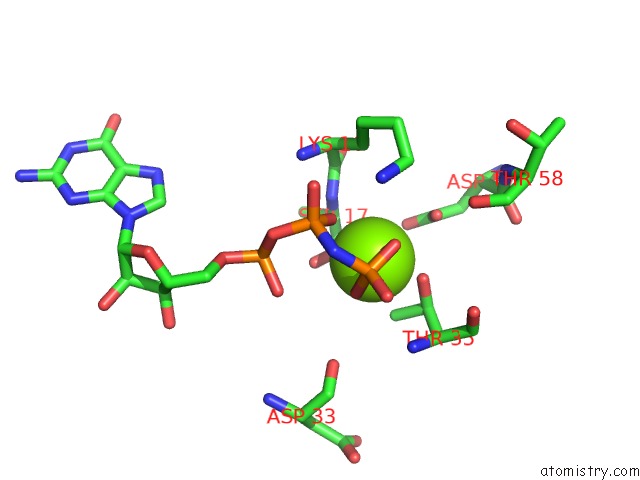
Mono view
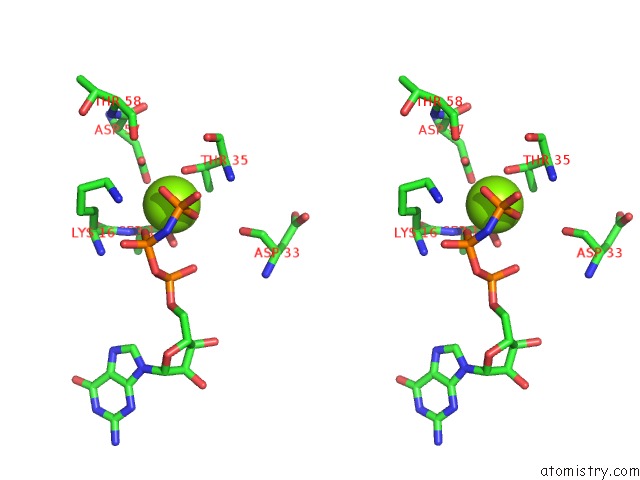
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of G12V Hras-Gppnhp Bound in Complex with the Engineered Rbd Variant 12 of Craf Kinase Protein within 5.0Å range:
|
Reference:
S.Wiechmann,
P.Maisonneuve,
B.M.Grebbin,
M.Hoffmeister,
M.Kaulich,
H.Clevers,
K.Rajalingam,
I.Kurinov,
H.F.Farin,
F.Sicheri,
A.Ernst.
Conformation-Specific Inhibitors of Activated Ras Gtpases Reveal Limited Ras Dependency of Patient-Derived Cancer Organoids. J.Biol.Chem. 2020.
ISSN: ESSN 1083-351X
PubMed: 32086379
DOI: 10.1074/JBC.RA119.011025
Page generated: Tue Oct 1 12:53:34 2024
ISSN: ESSN 1083-351X
PubMed: 32086379
DOI: 10.1074/JBC.RA119.011025
Last articles
Fe in 9FSRFe in 9FSQ
Fe in 9FSS
Fe in 9FSP
Fe in 9FSO
Fe in 9FMU
Fe in 9FHB
Fe in 9FBK
Fe in 9EBR
Fe in 9CU2