Magnesium »
PDB 6peu-6psq »
6pmo »
Magnesium in PDB 6pmo: Co-Crystal Structure of the Geobacillus Kaustophilus Glyq T-Box Riboswitch Discriminator Domain in Complex with Trna-Gly
Protein crystallography data
The structure of Co-Crystal Structure of the Geobacillus Kaustophilus Glyq T-Box Riboswitch Discriminator Domain in Complex with Trna-Gly, PDB code: 6pmo
was solved by
S.Li,
J.Zhang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 34.98 / 2.66 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 44.851, 139.913, 136.023, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 22.5 / 27.3 |
Other elements in 6pmo:
The structure of Co-Crystal Structure of the Geobacillus Kaustophilus Glyq T-Box Riboswitch Discriminator Domain in Complex with Trna-Gly also contains other interesting chemical elements:
| Iridium | (Ir) | 13 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Co-Crystal Structure of the Geobacillus Kaustophilus Glyq T-Box Riboswitch Discriminator Domain in Complex with Trna-Gly
(pdb code 6pmo). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Co-Crystal Structure of the Geobacillus Kaustophilus Glyq T-Box Riboswitch Discriminator Domain in Complex with Trna-Gly, PDB code: 6pmo:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Co-Crystal Structure of the Geobacillus Kaustophilus Glyq T-Box Riboswitch Discriminator Domain in Complex with Trna-Gly, PDB code: 6pmo:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 6pmo
Go back to
Magnesium binding site 1 out
of 4 in the Co-Crystal Structure of the Geobacillus Kaustophilus Glyq T-Box Riboswitch Discriminator Domain in Complex with Trna-Gly
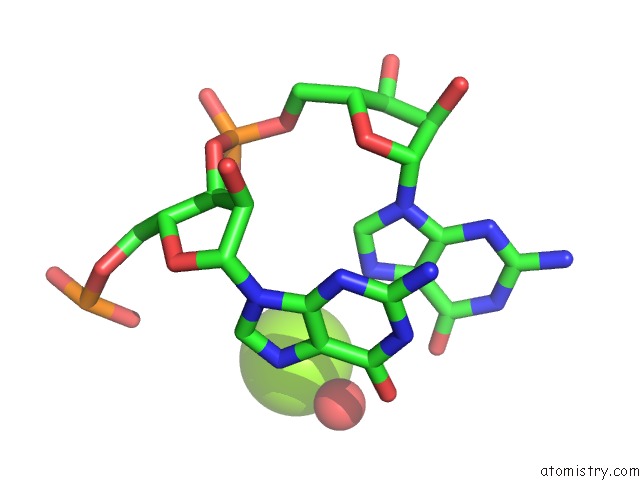
Mono view
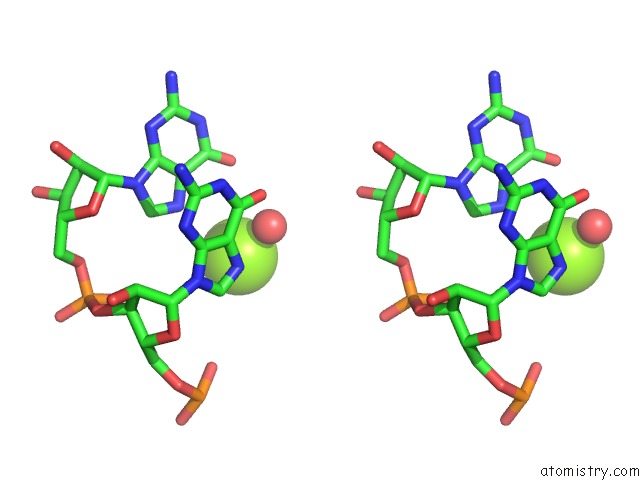
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Co-Crystal Structure of the Geobacillus Kaustophilus Glyq T-Box Riboswitch Discriminator Domain in Complex with Trna-Gly within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 6pmo
Go back to
Magnesium binding site 2 out
of 4 in the Co-Crystal Structure of the Geobacillus Kaustophilus Glyq T-Box Riboswitch Discriminator Domain in Complex with Trna-Gly
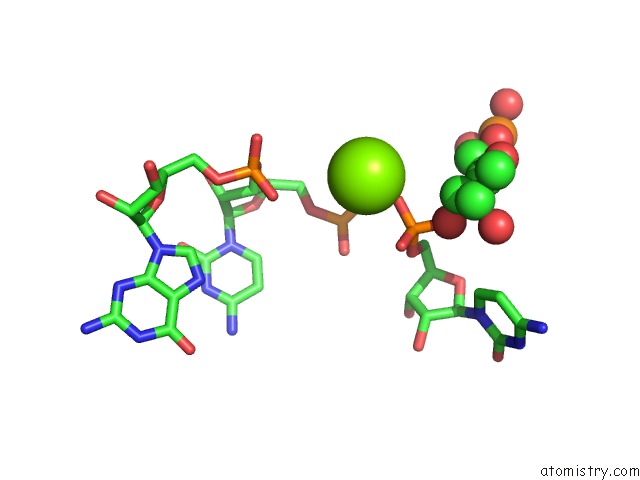
Mono view
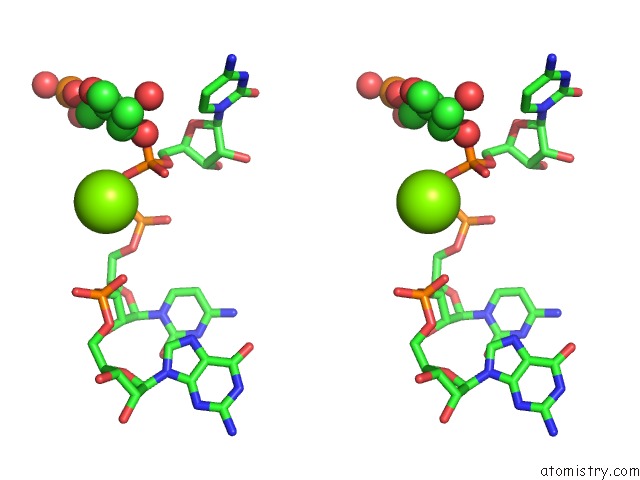
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Co-Crystal Structure of the Geobacillus Kaustophilus Glyq T-Box Riboswitch Discriminator Domain in Complex with Trna-Gly within 5.0Å range:
|
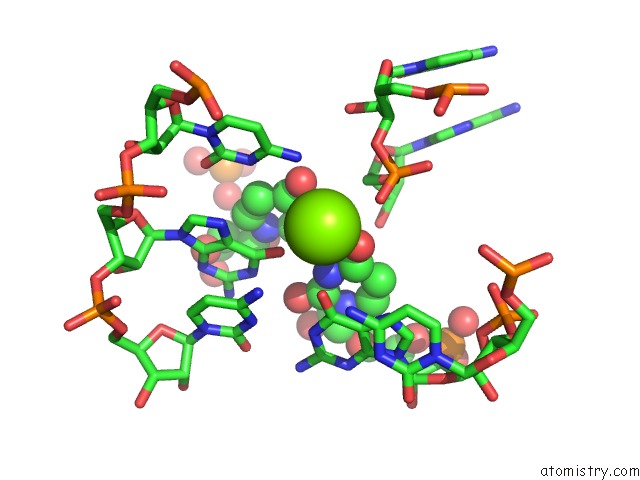
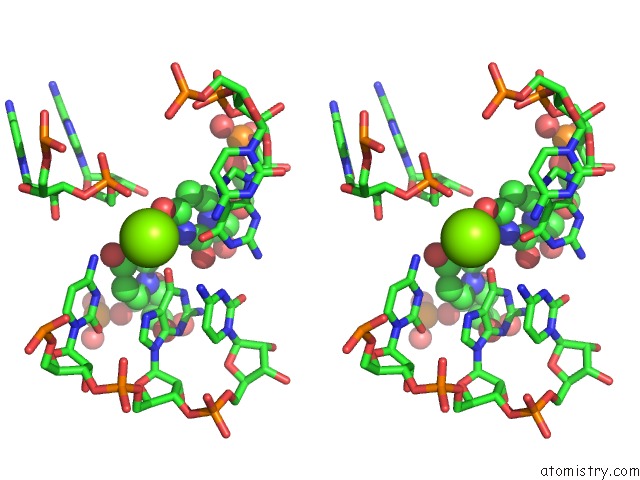
Magnesium binding site 3 out of 4 in 6pmo
Go back to
Magnesium binding site 3 out
of 4 in the Co-Crystal Structure of the Geobacillus Kaustophilus Glyq T-Box Riboswitch Discriminator Domain in Complex with Trna-Gly
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Co-Crystal Structure of the Geobacillus Kaustophilus Glyq T-Box Riboswitch Discriminator Domain in Complex with Trna-Gly within 5.0Å range:
|
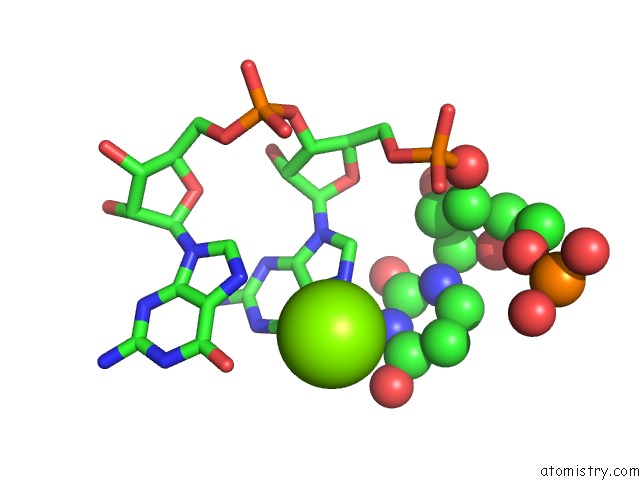
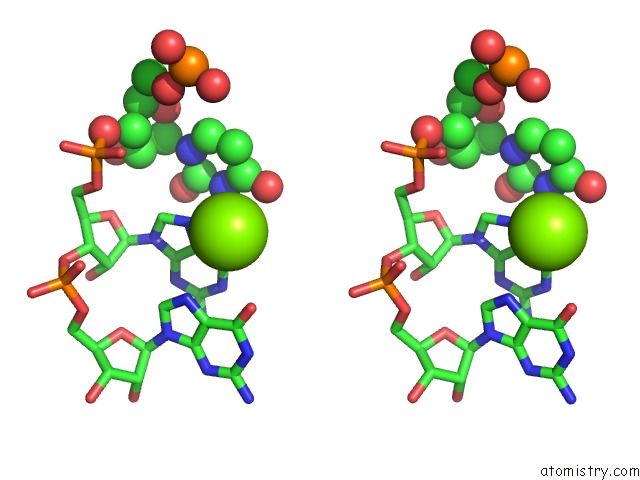
Magnesium binding site 4 out of 4 in 6pmo
Go back to
Magnesium binding site 4 out
of 4 in the Co-Crystal Structure of the Geobacillus Kaustophilus Glyq T-Box Riboswitch Discriminator Domain in Complex with Trna-Gly
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Co-Crystal Structure of the Geobacillus Kaustophilus Glyq T-Box Riboswitch Discriminator Domain in Complex with Trna-Gly within 5.0Å range:
|
Reference:
S.Li,
Z.Su,
J.Lehmann,
V.Stamatopoulou,
N.Giarimoglou,
F.E.Henderson,
L.Fan,
G.D.Pintilie,
K.Zhang,
M.Chen,
S.J.Ludtke,
Y.X.Wang,
C.Stathopoulos,
W.Chiu,
J.Zhang.
Structural Basis of Amino Acid Surveillance By Higher-Order Trna-Mrna Interactions. Nat.Struct.Mol.Biol. 2019.
ISSN: ESSN 1545-9985
PubMed: 31740854
DOI: 10.1038/S41594-019-0326-7
Page generated: Tue Oct 1 14:13:21 2024
ISSN: ESSN 1545-9985
PubMed: 31740854
DOI: 10.1038/S41594-019-0326-7
Last articles
Cl in 7TNLCl in 7TNK
Cl in 7TNJ
Cl in 7TNH
Cl in 7TNF
Cl in 7TND
Cl in 7TN8
Cl in 7TNC
Cl in 7TN7
Cl in 7TN4