Magnesium »
PDB 6quz-6r5l »
6r3q »
Magnesium in PDB 6r3q: The Structure of A Membrane Adenylyl Cyclase Bound to An Activated Stimulatory G Protein
Magnesium Binding Sites:
The binding sites of Magnesium atom in the The Structure of A Membrane Adenylyl Cyclase Bound to An Activated Stimulatory G Protein
(pdb code 6r3q). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the The Structure of A Membrane Adenylyl Cyclase Bound to An Activated Stimulatory G Protein, PDB code: 6r3q:
In total only one binding site of Magnesium was determined in the The Structure of A Membrane Adenylyl Cyclase Bound to An Activated Stimulatory G Protein, PDB code: 6r3q:
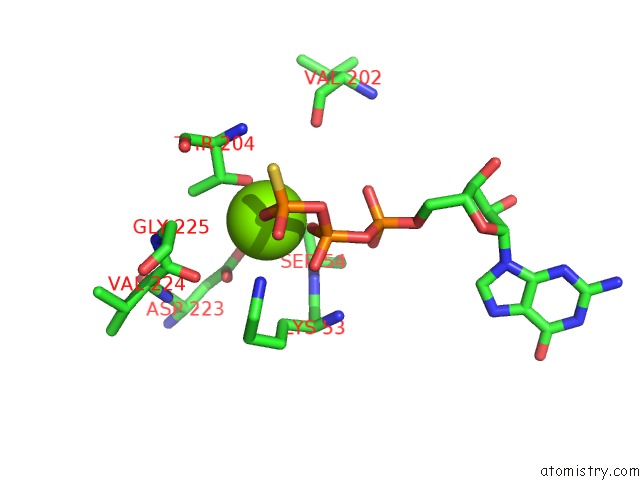
Magnesium binding site 1 out of 1 in 6r3q
Go back to
Magnesium binding site 1 out
of 1 in the The Structure of A Membrane Adenylyl Cyclase Bound to An Activated Stimulatory G Protein
Mono view
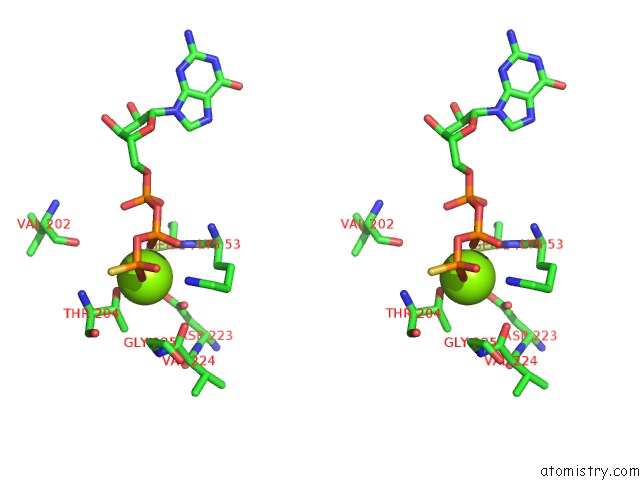
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of The Structure of A Membrane Adenylyl Cyclase Bound to An Activated Stimulatory G Protein within 5.0Å range:
|
Reference:
C.Qi,
S.Sorrentino,
O.Medalia,
V.M.Korkhov.
The Structure of A Membrane Adenylyl Cyclase Bound to An Activated Stimulatory G Protein. Science V. 364 389 2019.
ISSN: ESSN 1095-9203
PubMed: 31023924
DOI: 10.1126/SCIENCE.AAV0778
Page generated: Tue Oct 1 16:32:30 2024
ISSN: ESSN 1095-9203
PubMed: 31023924
DOI: 10.1126/SCIENCE.AAV0778
Last articles
F in 7LG8F in 7LD3
F in 7LCR
F in 7LCM
F in 7LCO
F in 7LCK
F in 7LCJ
F in 7LCI
F in 7L9Y
F in 7LCD