Magnesium »
PDB 6rdk-6rf8 »
6rdx »
Magnesium in PDB 6rdx: Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement
Enzymatic activity of Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement
All present enzymatic activity of Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement:
7.1.2.2;
7.1.2.2;
Other elements in 6rdx:
The structure of Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement
(pdb code 6rdx). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 5 binding sites of Magnesium where determined in the Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement, PDB code: 6rdx:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Magnesium where determined in the Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement, PDB code: 6rdx:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5;
Magnesium binding site 1 out of 5 in 6rdx
Go back to
Magnesium binding site 1 out
of 5 in the Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement
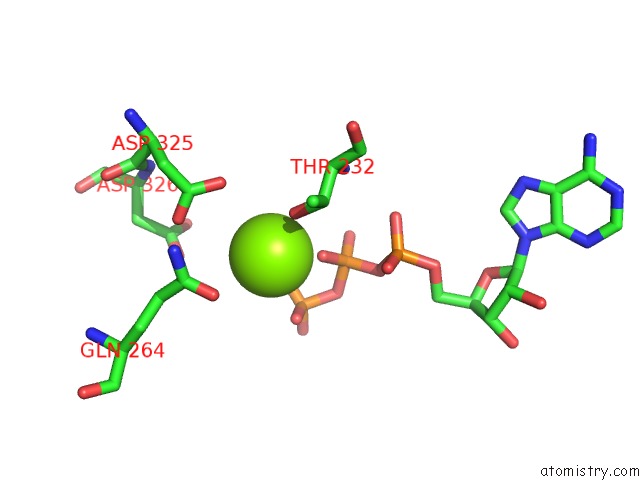
Mono view
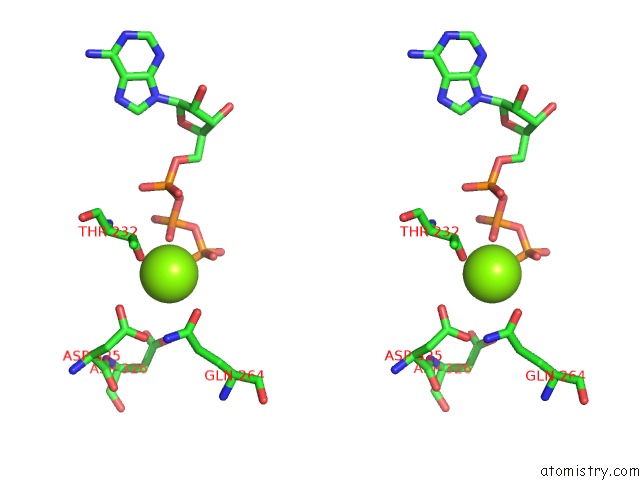
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement within 5.0Å range:
|
Magnesium binding site 2 out of 5 in 6rdx
Go back to
Magnesium binding site 2 out
of 5 in the Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement
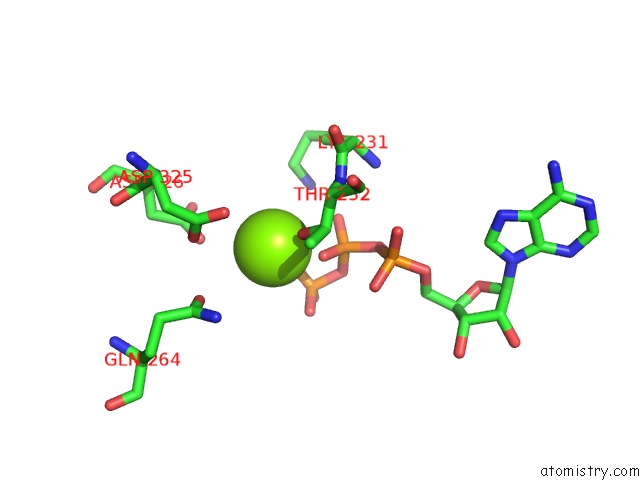
Mono view
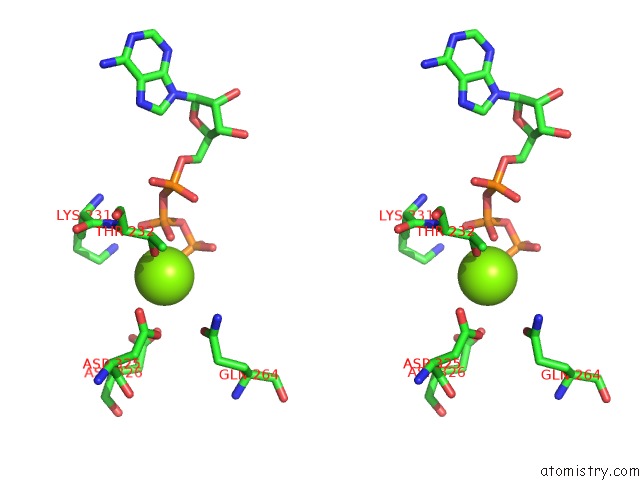
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement within 5.0Å range:
|
Magnesium binding site 3 out of 5 in 6rdx
Go back to
Magnesium binding site 3 out
of 5 in the Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement
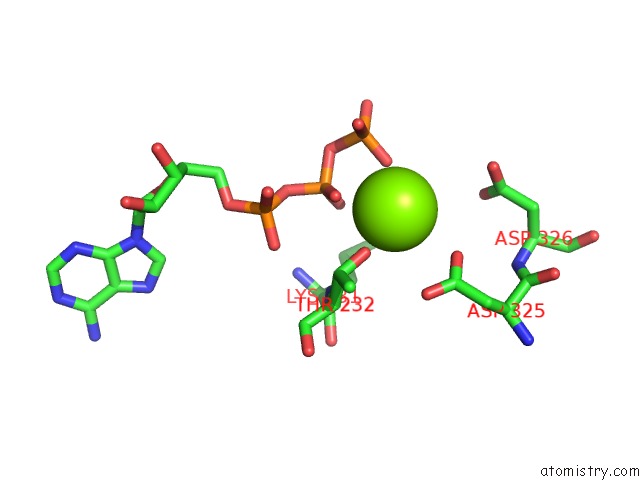
Mono view
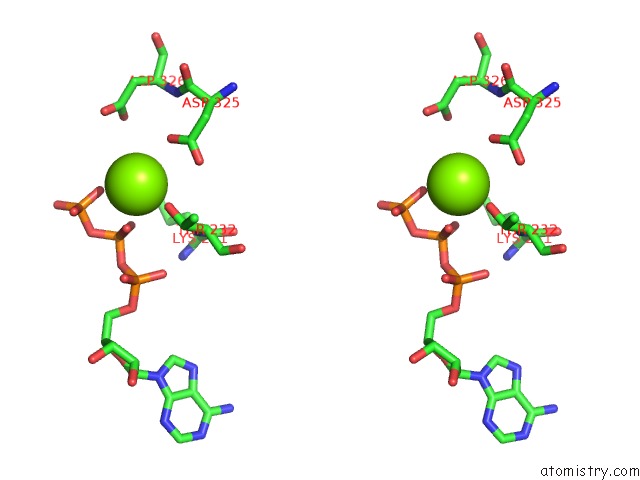
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement within 5.0Å range:
|
Magnesium binding site 4 out of 5 in 6rdx
Go back to
Magnesium binding site 4 out
of 5 in the Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement within 5.0Å range:
|
Magnesium binding site 5 out of 5 in 6rdx
Go back to
Magnesium binding site 5 out
of 5 in the Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement
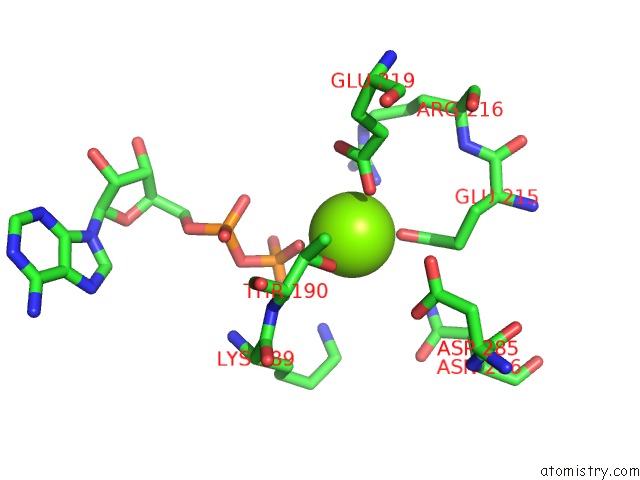
Mono view
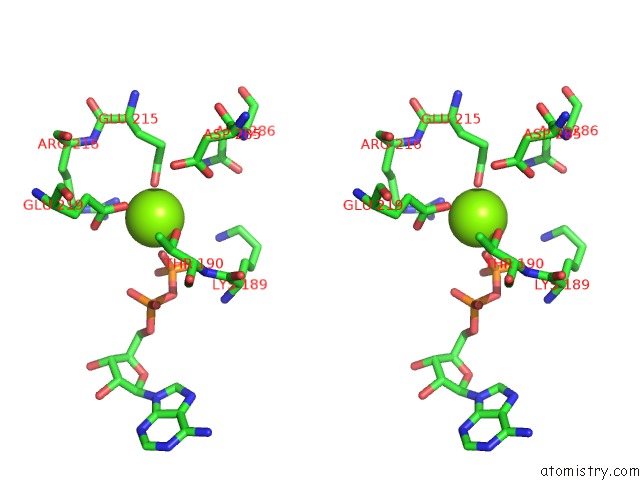
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Cryo-Em Structure of Polytomella F-Atp Synthase, Rotary Substate 1F, Monomer-Masked Refinement within 5.0Å range:
|
Reference:
B.J.Murphy,
N.Klusch,
J.Langer,
D.J.Mills,
O.Yildiz,
W.Kuhlbrandt.
Rotary Substates of Mitochondrial Atp Synthase Reveal the Basis of Flexible F1-Focoupling. Science V. 364 2019.
ISSN: ESSN 1095-9203
PubMed: 31221832
DOI: 10.1126/SCIENCE.AAW9128
Page generated: Wed Aug 13 15:39:14 2025
ISSN: ESSN 1095-9203
PubMed: 31221832
DOI: 10.1126/SCIENCE.AAW9128
Last articles
Mg in 7V3MMg in 7V3C
Mg in 7V3B
Mg in 7V3A
Mg in 7V33
Mg in 7V32
Mg in 7V31
Mg in 7V30
Mg in 7V2R
Mg in 7V2K