Magnesium »
PDB 6tsh-6u1k »
6ty9 »
Magnesium in PDB 6ty9: In Situ Structure of Bmcpv Rna Dependent Rna Polymerase at Initiation State
Magnesium Binding Sites:
The binding sites of Magnesium atom in the In Situ Structure of Bmcpv Rna Dependent Rna Polymerase at Initiation State
(pdb code 6ty9). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the In Situ Structure of Bmcpv Rna Dependent Rna Polymerase at Initiation State, PDB code: 6ty9:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the In Situ Structure of Bmcpv Rna Dependent Rna Polymerase at Initiation State, PDB code: 6ty9:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 6ty9
Go back to
Magnesium binding site 1 out
of 3 in the In Situ Structure of Bmcpv Rna Dependent Rna Polymerase at Initiation State
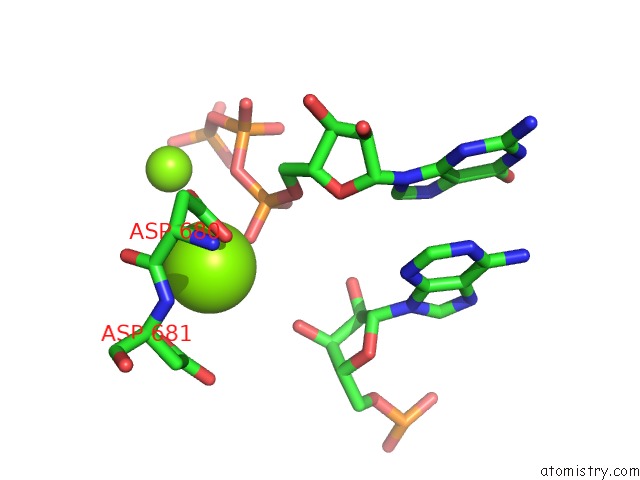
Mono view
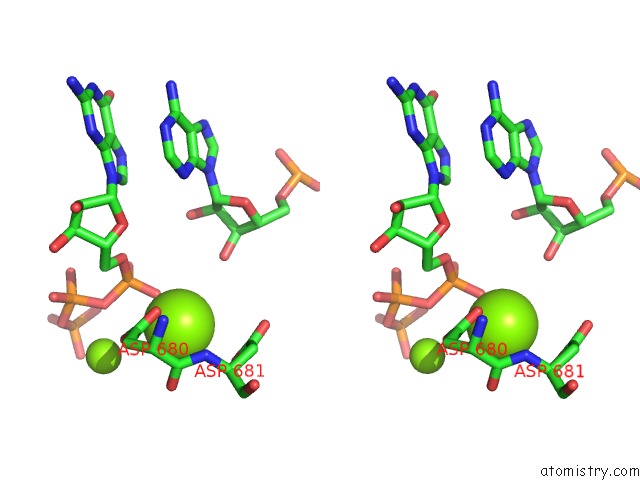
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of In Situ Structure of Bmcpv Rna Dependent Rna Polymerase at Initiation State within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 6ty9
Go back to
Magnesium binding site 2 out
of 3 in the In Situ Structure of Bmcpv Rna Dependent Rna Polymerase at Initiation State
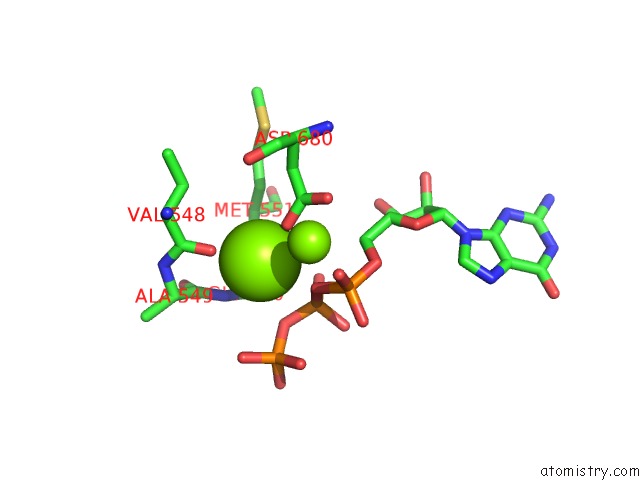
Mono view
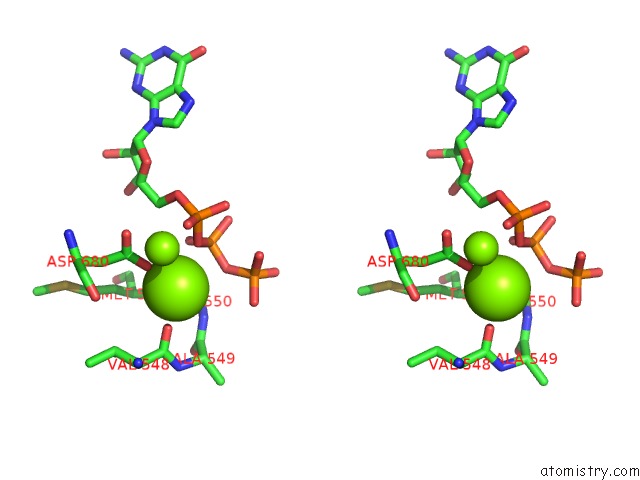
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of In Situ Structure of Bmcpv Rna Dependent Rna Polymerase at Initiation State within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 6ty9
Go back to
Magnesium binding site 3 out
of 3 in the In Situ Structure of Bmcpv Rna Dependent Rna Polymerase at Initiation State
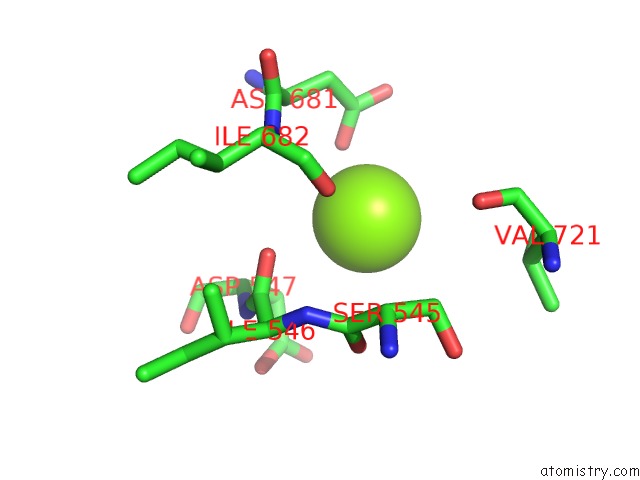
Mono view
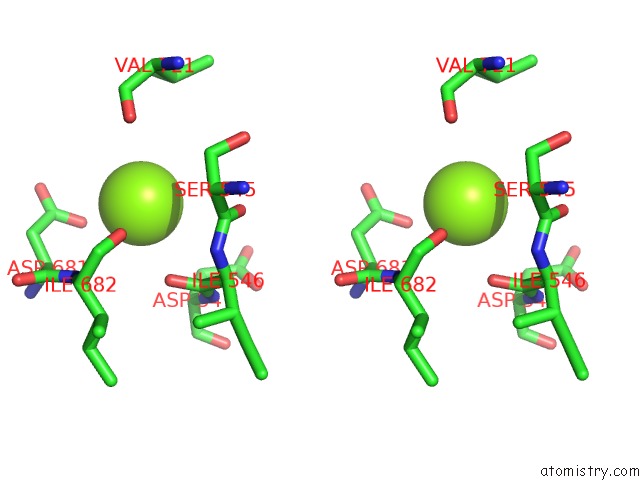
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of In Situ Structure of Bmcpv Rna Dependent Rna Polymerase at Initiation State within 5.0Å range:
|
Reference:
Y.Cui,
Y.Zhang,
K.Zhou,
J.Sun,
Z.H.Zhou.
Conservative Transcription in Three Steps Visualized in A Double-Stranded Rna Virus. Nat.Struct.Mol.Biol. V. 26 1023 2019.
ISSN: ESSN 1545-9985
PubMed: 31695188
DOI: 10.1038/S41594-019-0320-0
Page generated: Tue Oct 1 20:35:00 2024
ISSN: ESSN 1545-9985
PubMed: 31695188
DOI: 10.1038/S41594-019-0320-0
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO