Magnesium »
PDB 6ufj-6up4 »
6umq »
Magnesium in PDB 6umq: Structure of DUF89
Protein crystallography data
The structure of Structure of DUF89, PDB code: 6umq
was solved by
J.J.Perry,
N.Kenjic,
T.N.Dennis,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 66.49 / 1.85 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 90.140, 194.473, 114.201, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.6 / 22.4 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of DUF89
(pdb code 6umq). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Structure of DUF89, PDB code: 6umq:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Structure of DUF89, PDB code: 6umq:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 6umq
Go back to
Magnesium binding site 1 out
of 4 in the Structure of DUF89
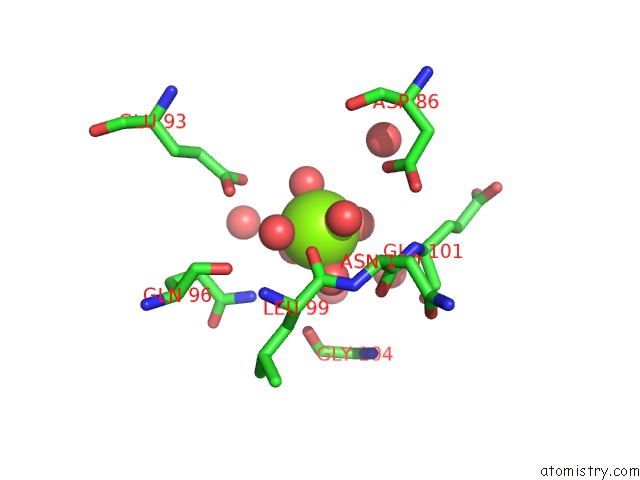
Mono view
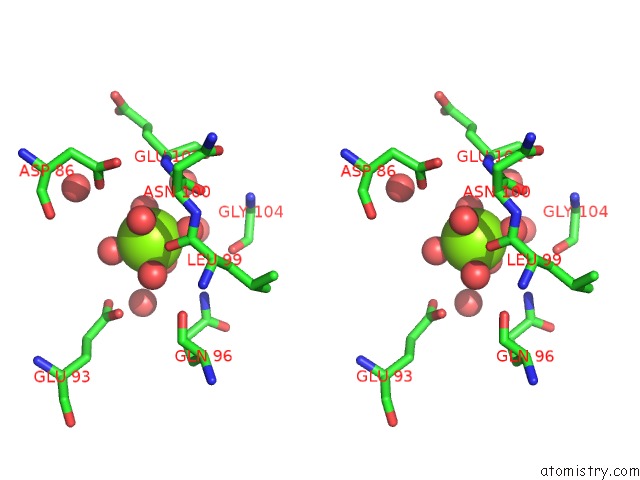
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of DUF89 within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 6umq
Go back to
Magnesium binding site 2 out
of 4 in the Structure of DUF89
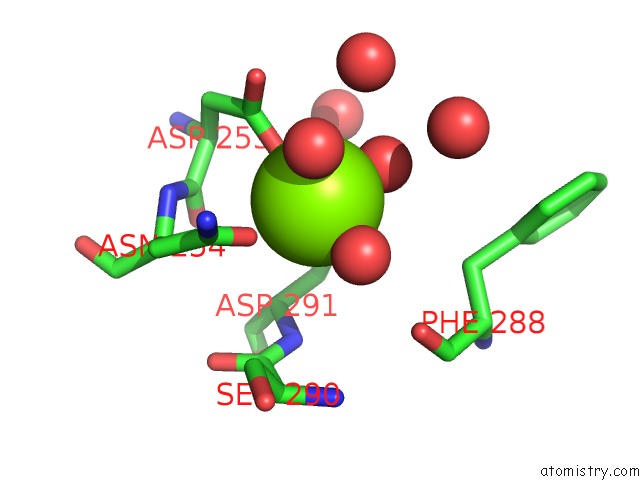
Mono view
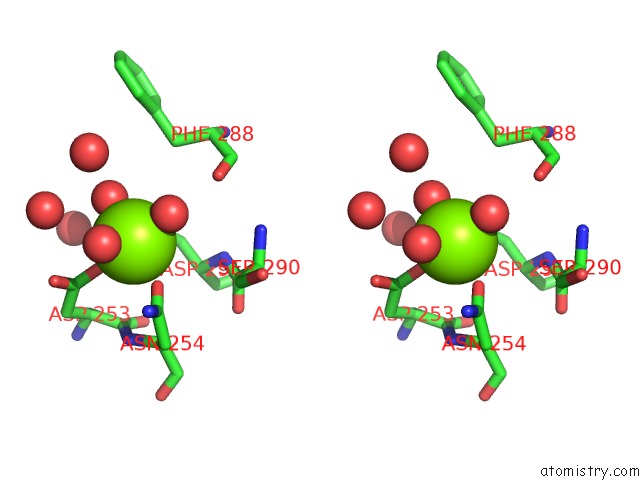
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of DUF89 within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 6umq
Go back to
Magnesium binding site 3 out
of 4 in the Structure of DUF89
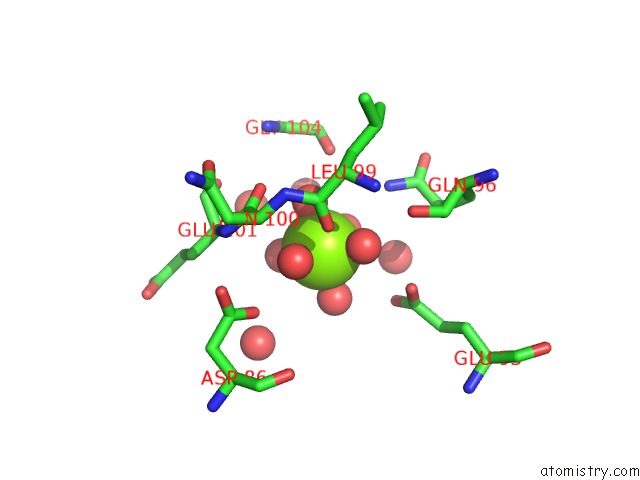
Mono view
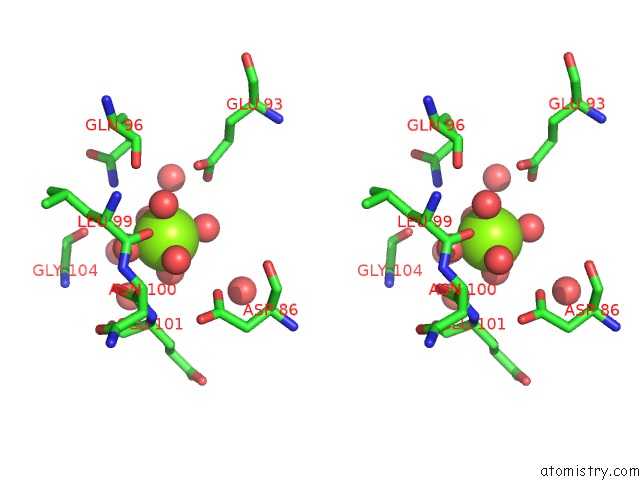
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structure of DUF89 within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 6umq
Go back to
Magnesium binding site 4 out
of 4 in the Structure of DUF89
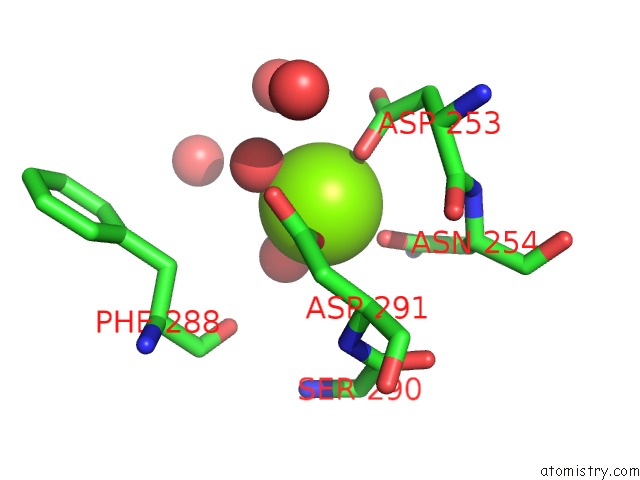
Mono view
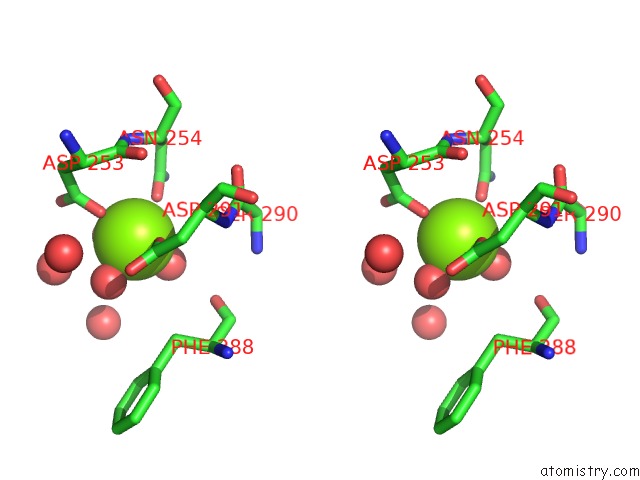
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Structure of DUF89 within 5.0Å range:
|
Reference:
T.N.Dennis,
N.Kenjic,
A.S.Kang,
J.D.Lowenson,
J.S.Kirkwood,
S.G.Clarke,
J.J.P.Perry.
Human ARMT1 Structure and Substrate Specificity Indicates That It Is A DUF89 Family Damage-Control Phosphatase. J.Struct.Biol. V. 212 07576 2020.
ISSN: ESSN 1095-8657
PubMed: 32682077
DOI: 10.1016/J.JSB.2020.107576
Page generated: Tue Oct 1 21:07:24 2024
ISSN: ESSN 1095-8657
PubMed: 32682077
DOI: 10.1016/J.JSB.2020.107576
Last articles
Cl in 6BB0Cl in 6BAX
Cl in 6BAZ
Cl in 6BAR
Cl in 6BAS
Cl in 6BAQ
Cl in 6BAG
Cl in 6BAD
Cl in 6BA0
Cl in 6BAA