Magnesium »
PDB 6upd-6uzk »
6ut6 »
Magnesium in PDB 6ut6: Cryo-Em Structure of the Escherichia Coli Mcrbc Complex
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of the Escherichia Coli Mcrbc Complex
(pdb code 6ut6). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Cryo-Em Structure of the Escherichia Coli Mcrbc Complex, PDB code: 6ut6:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Cryo-Em Structure of the Escherichia Coli Mcrbc Complex, PDB code: 6ut6:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 6ut6
Go back to
Magnesium binding site 1 out
of 4 in the Cryo-Em Structure of the Escherichia Coli Mcrbc Complex
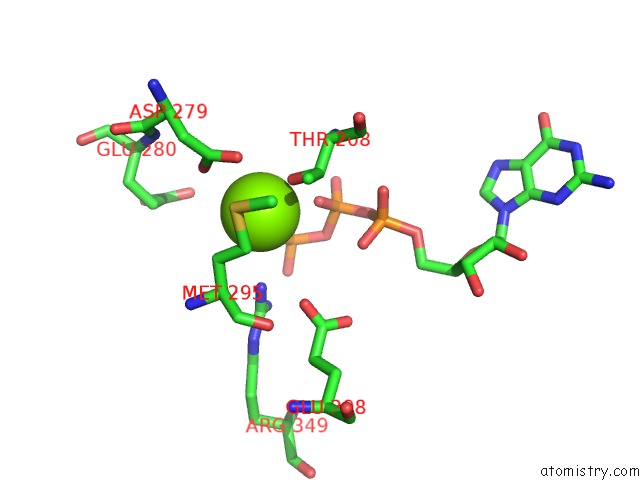
Mono view
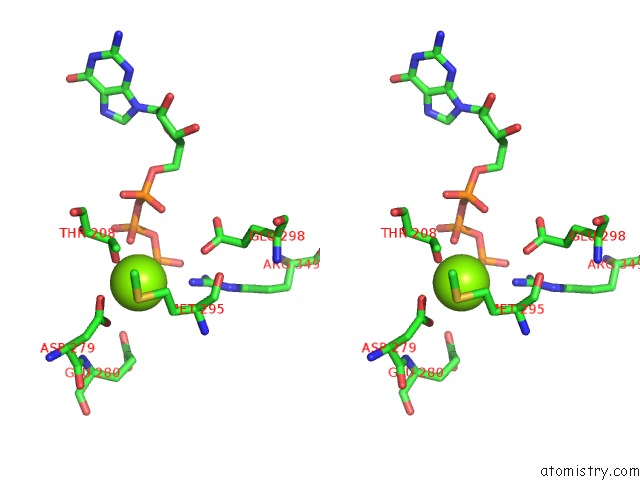
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of the Escherichia Coli Mcrbc Complex within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 6ut6
Go back to
Magnesium binding site 2 out
of 4 in the Cryo-Em Structure of the Escherichia Coli Mcrbc Complex
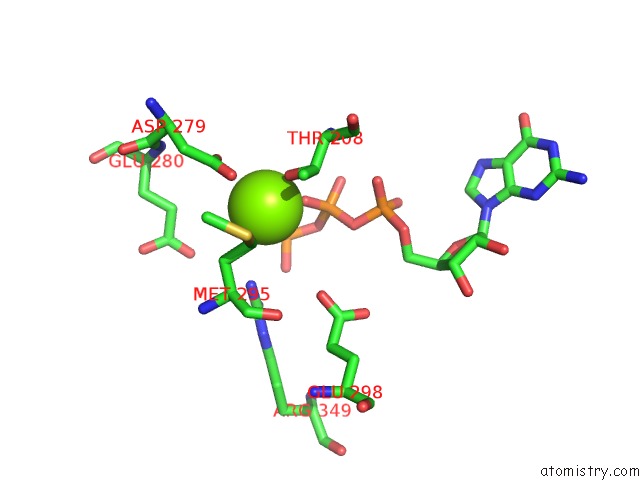
Mono view
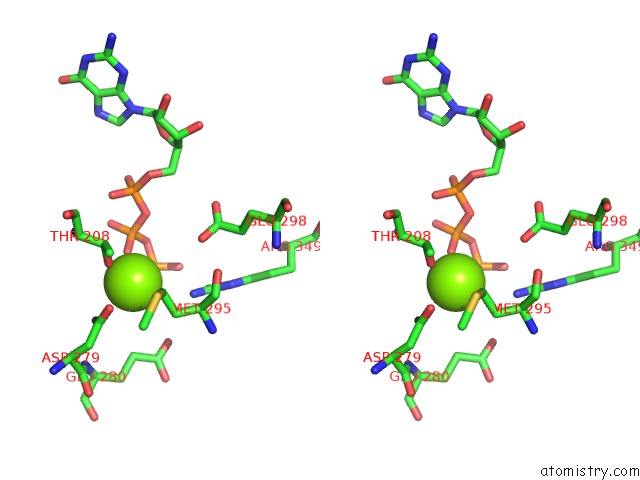
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Cryo-Em Structure of the Escherichia Coli Mcrbc Complex within 5.0Å range:
|
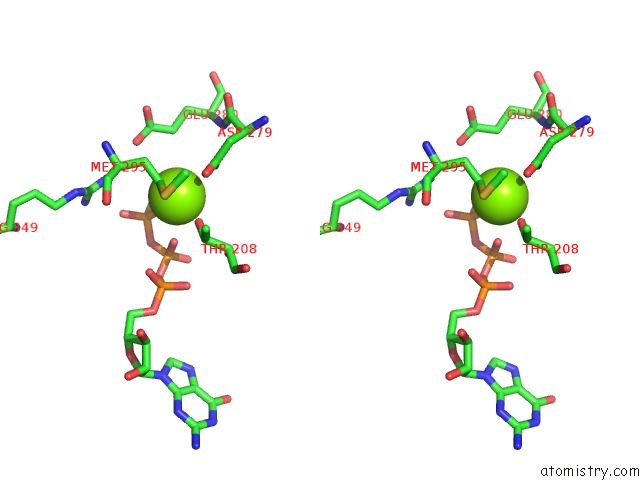
Magnesium binding site 3 out of 4 in 6ut6
Go back to
Magnesium binding site 3 out
of 4 in the Cryo-Em Structure of the Escherichia Coli Mcrbc Complex
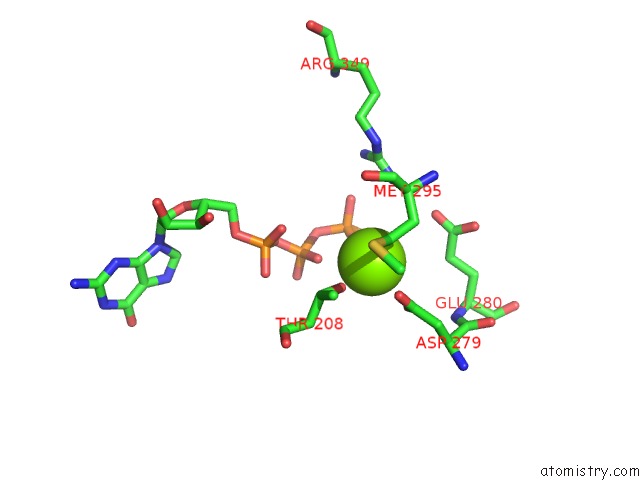
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Cryo-Em Structure of the Escherichia Coli Mcrbc Complex within 5.0Å range:
|
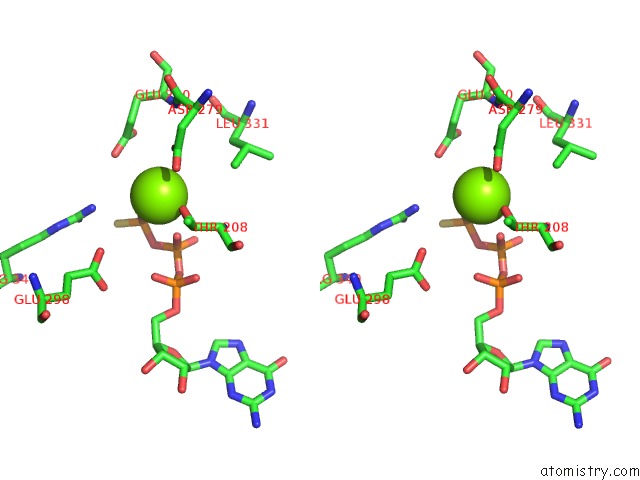
Magnesium binding site 4 out of 4 in 6ut6
Go back to
Magnesium binding site 4 out
of 4 in the Cryo-Em Structure of the Escherichia Coli Mcrbc Complex
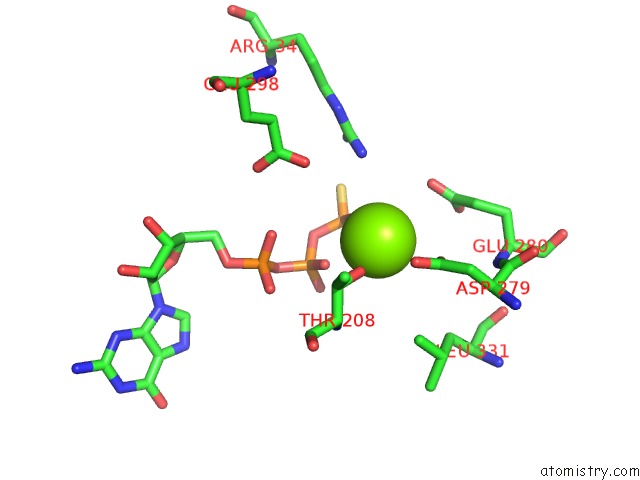
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Cryo-Em Structure of the Escherichia Coli Mcrbc Complex within 5.0Å range:
|
Reference:
Y.Niu,
H.Suzuki,
C.J.Hosford,
T.Walz,
J.S.Chappie.
Structural Asymmetry Governs the Assembly and Gtpase Activity of Mcrbc Restriction Complexes To Be Published.
Page generated: Wed Aug 13 18:43:20 2025
Last articles
Mg in 7KXGMg in 7KZ2
Mg in 7KZ4
Mg in 7KYB
Mg in 7KYC
Mg in 7KYA
Mg in 7KXH
Mg in 7KY9
Mg in 7KY8
Mg in 7KY7