Magnesium »
PDB 6vcw-6vo2 »
6vfx »
Magnesium in PDB 6vfx: Clpxp From Neisseria Meningitidis - Conformation B
Enzymatic activity of Clpxp From Neisseria Meningitidis - Conformation B
All present enzymatic activity of Clpxp From Neisseria Meningitidis - Conformation B:
3.4.21.92;
3.4.21.92;
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Clpxp From Neisseria Meningitidis - Conformation B
(pdb code 6vfx). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 5 binding sites of Magnesium where determined in the Clpxp From Neisseria Meningitidis - Conformation B, PDB code: 6vfx:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Magnesium where determined in the Clpxp From Neisseria Meningitidis - Conformation B, PDB code: 6vfx:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5;
Magnesium binding site 1 out of 5 in 6vfx
Go back to
Magnesium binding site 1 out
of 5 in the Clpxp From Neisseria Meningitidis - Conformation B
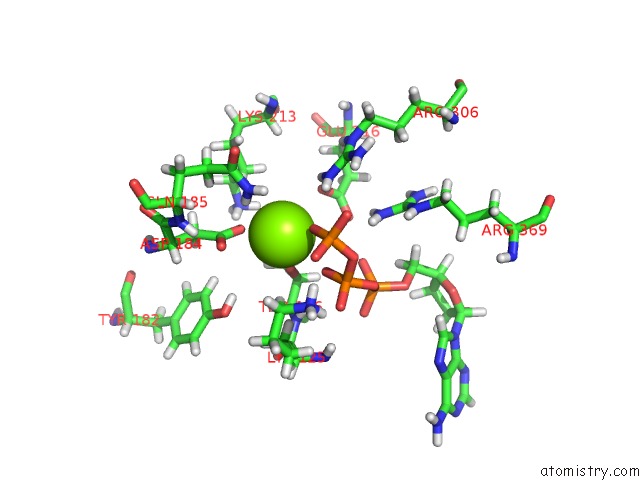
Mono view
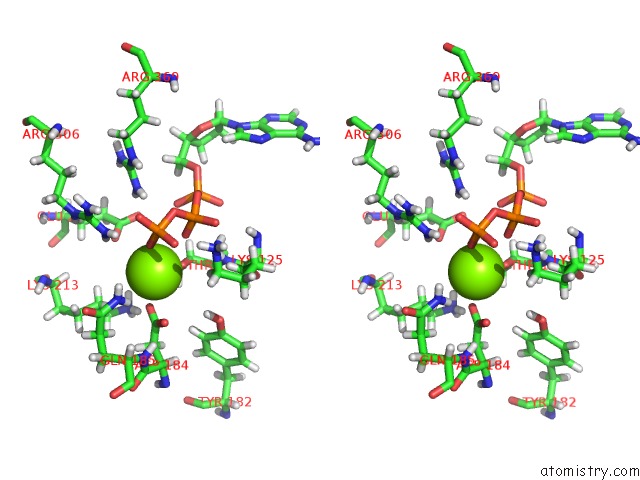
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Clpxp From Neisseria Meningitidis - Conformation B within 5.0Å range:
|
Magnesium binding site 2 out of 5 in 6vfx
Go back to
Magnesium binding site 2 out
of 5 in the Clpxp From Neisseria Meningitidis - Conformation B
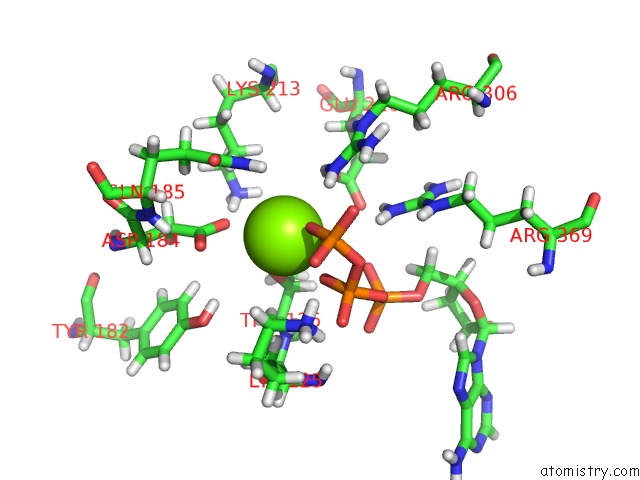
Mono view
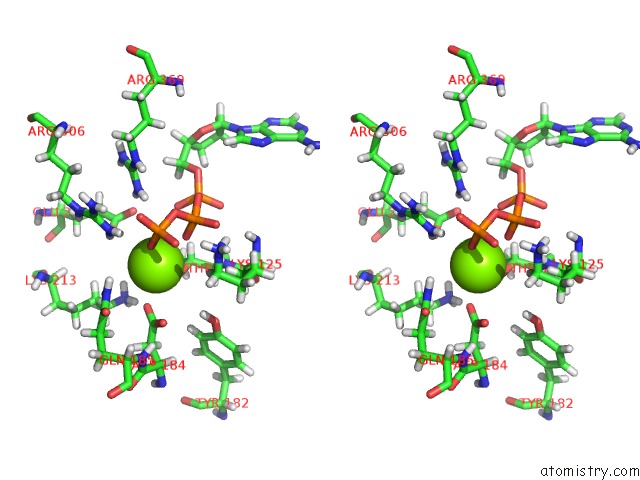
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Clpxp From Neisseria Meningitidis - Conformation B within 5.0Å range:
|
Magnesium binding site 3 out of 5 in 6vfx
Go back to
Magnesium binding site 3 out
of 5 in the Clpxp From Neisseria Meningitidis - Conformation B
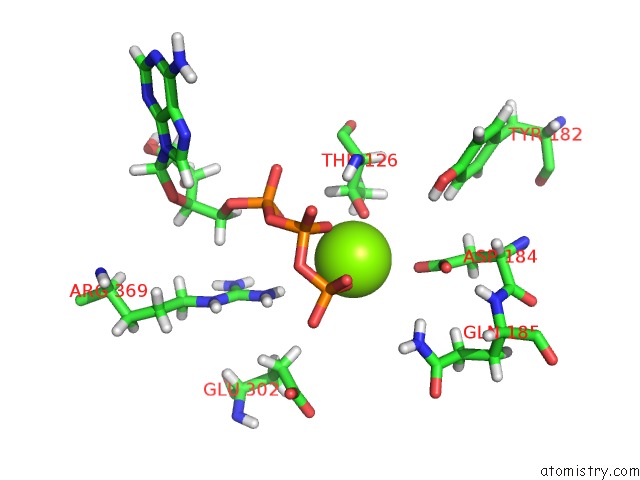
Mono view
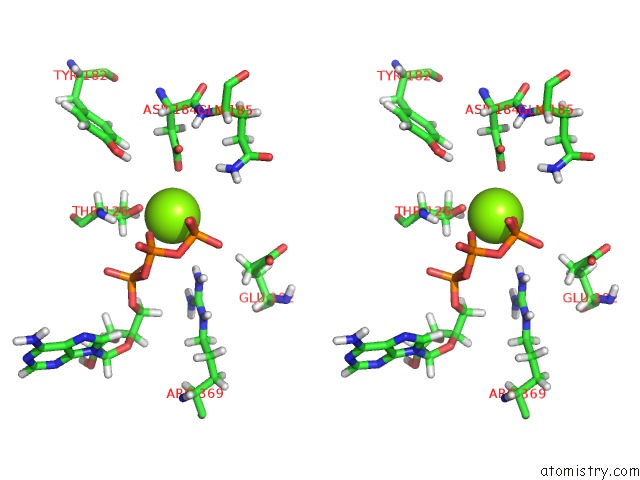
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Clpxp From Neisseria Meningitidis - Conformation B within 5.0Å range:
|
Magnesium binding site 4 out of 5 in 6vfx
Go back to
Magnesium binding site 4 out
of 5 in the Clpxp From Neisseria Meningitidis - Conformation B
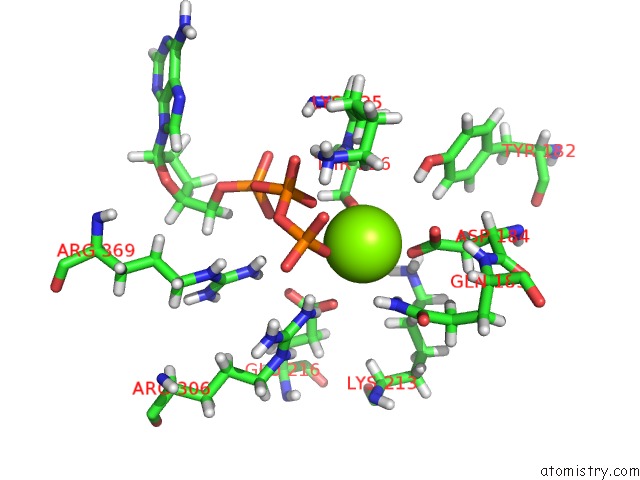
Mono view
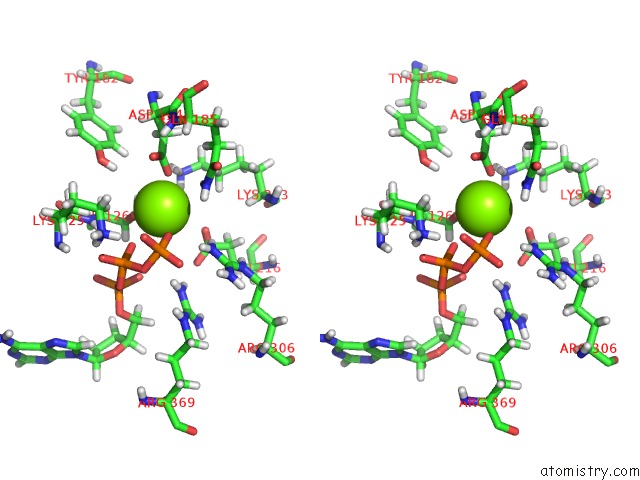
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Clpxp From Neisseria Meningitidis - Conformation B within 5.0Å range:
|
Magnesium binding site 5 out of 5 in 6vfx
Go back to
Magnesium binding site 5 out
of 5 in the Clpxp From Neisseria Meningitidis - Conformation B
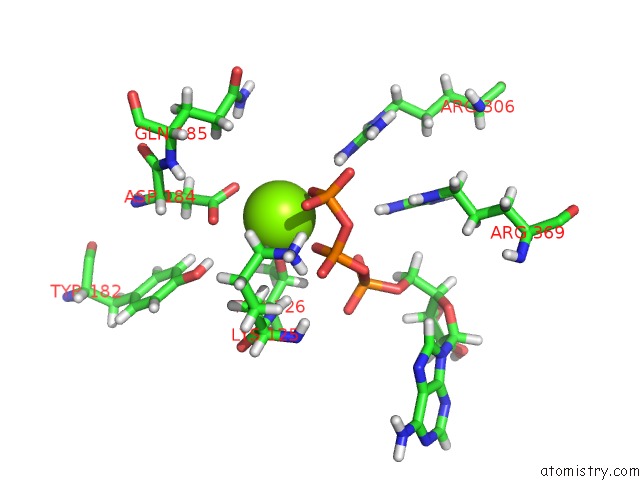
Mono view
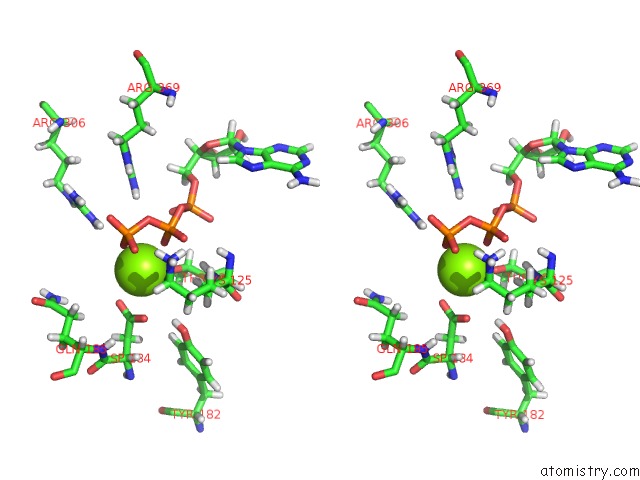
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Clpxp From Neisseria Meningitidis - Conformation B within 5.0Å range:
|
Reference:
Z.A.Ripstein,
S.Vahidi,
W.A.Houry,
J.L.Rubinstein,
L.E.Kay.
A Processive Rotary Mechanism Couples Substrate Unfolding and Proteolysis in the Clpxp Degradation Machinery. Elife V. 9 2020.
ISSN: ESSN 2050-084X
PubMed: 31916936
DOI: 10.7554/ELIFE.52158
Page generated: Tue Oct 1 22:03:56 2024
ISSN: ESSN 2050-084X
PubMed: 31916936
DOI: 10.7554/ELIFE.52158
Last articles
Cl in 7TMUCl in 7TJP
Cl in 7TMZ
Cl in 7TLG
Cl in 7TJC
Cl in 7TJO
Cl in 7TLE
Cl in 7TKV
Cl in 7THH
Cl in 7TIV