Magnesium »
PDB 6vzs-6wic »
6w2l »
Magnesium in PDB 6w2l: Crystal Structure of Human Dehydrodolichyl Diphosphate Synthase (Ngbr/Dhdds) in Complex with Mg and Ipp
Enzymatic activity of Crystal Structure of Human Dehydrodolichyl Diphosphate Synthase (Ngbr/Dhdds) in Complex with Mg and Ipp
All present enzymatic activity of Crystal Structure of Human Dehydrodolichyl Diphosphate Synthase (Ngbr/Dhdds) in Complex with Mg and Ipp:
2.5.1.87;
2.5.1.87;
Protein crystallography data
The structure of Crystal Structure of Human Dehydrodolichyl Diphosphate Synthase (Ngbr/Dhdds) in Complex with Mg and Ipp, PDB code: 6w2l
was solved by
B.H.Edani,
Y.Ha,
W.C.Sessa,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.92 / 2.31 |
| Space group | H 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 185.670, 185.670, 113.351, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 21.2 / 25.5 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Human Dehydrodolichyl Diphosphate Synthase (Ngbr/Dhdds) in Complex with Mg and Ipp
(pdb code 6w2l). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of Human Dehydrodolichyl Diphosphate Synthase (Ngbr/Dhdds) in Complex with Mg and Ipp, PDB code: 6w2l:
In total only one binding site of Magnesium was determined in the Crystal Structure of Human Dehydrodolichyl Diphosphate Synthase (Ngbr/Dhdds) in Complex with Mg and Ipp, PDB code: 6w2l:
Magnesium binding site 1 out of 1 in 6w2l
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of Human Dehydrodolichyl Diphosphate Synthase (Ngbr/Dhdds) in Complex with Mg and Ipp
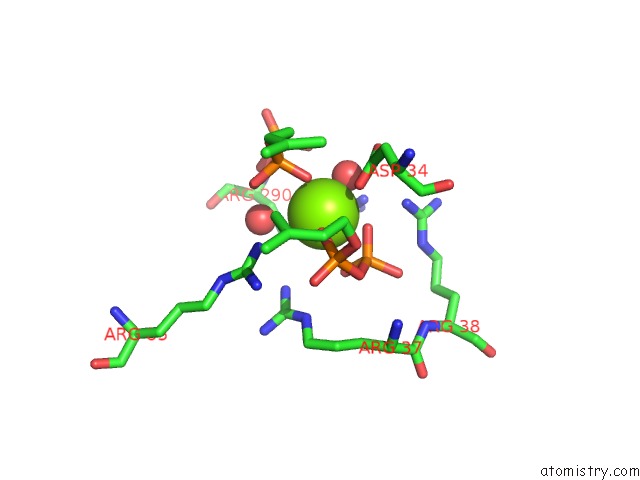
Mono view
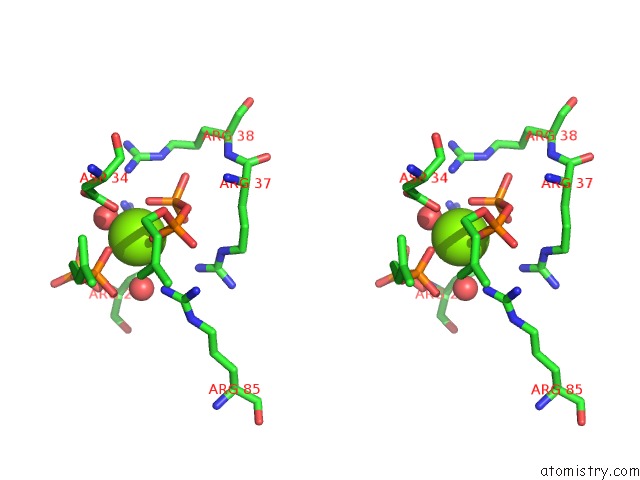
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Human Dehydrodolichyl Diphosphate Synthase (Ngbr/Dhdds) in Complex with Mg and Ipp within 5.0Å range:
|
Reference:
B.H.Edani,
K.A.Grabinska,
R.Zhang,
E.J.Park,
B.Siciliano,
L.Surmacz,
Y.Ha,
W.C.Sessa.
Structural Elucidation of Thecis-Prenyltransferase Ngbr/Dhdds Complex Reveals Insights in Regulation of Protein Glycosylation. Proc.Natl.Acad.Sci.Usa V. 117 20794 2020.
ISSN: ESSN 1091-6490
PubMed: 32817466
DOI: 10.1073/PNAS.2008381117
Page generated: Tue Oct 1 22:55:02 2024
ISSN: ESSN 1091-6490
PubMed: 32817466
DOI: 10.1073/PNAS.2008381117
Last articles
Cl in 7SUUCl in 7SUJ
Cl in 7SUT
Cl in 7SUI
Cl in 7SUH
Cl in 7SQE
Cl in 7STV
Cl in 7STT
Cl in 7STQ
Cl in 7SSF