Magnesium »
PDB 6wqg-6wwm »
6wtr »
Magnesium in PDB 6wtr: Structure of Human Pir-Mirna-300 Apical Loop Fused to the Ydao Riboswitch Scaffold
Protein crystallography data
The structure of Structure of Human Pir-Mirna-300 Apical Loop Fused to the Ydao Riboswitch Scaffold, PDB code: 6wtr
was solved by
G.M.Shoffner,
Z.Peng,
F.Guo,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 74.30 / 3.08 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 113.07, 113.07, 114.05, 90, 90, 120 |
| R / Rfree (%) | 16.5 / 19.2 |
Other elements in 6wtr:
The structure of Structure of Human Pir-Mirna-300 Apical Loop Fused to the Ydao Riboswitch Scaffold also contains other interesting chemical elements:
| Potassium | (K) | 1 atom |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of Human Pir-Mirna-300 Apical Loop Fused to the Ydao Riboswitch Scaffold
(pdb code 6wtr). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Structure of Human Pir-Mirna-300 Apical Loop Fused to the Ydao Riboswitch Scaffold, PDB code: 6wtr:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Structure of Human Pir-Mirna-300 Apical Loop Fused to the Ydao Riboswitch Scaffold, PDB code: 6wtr:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 6wtr
Go back to
Magnesium binding site 1 out
of 3 in the Structure of Human Pir-Mirna-300 Apical Loop Fused to the Ydao Riboswitch Scaffold
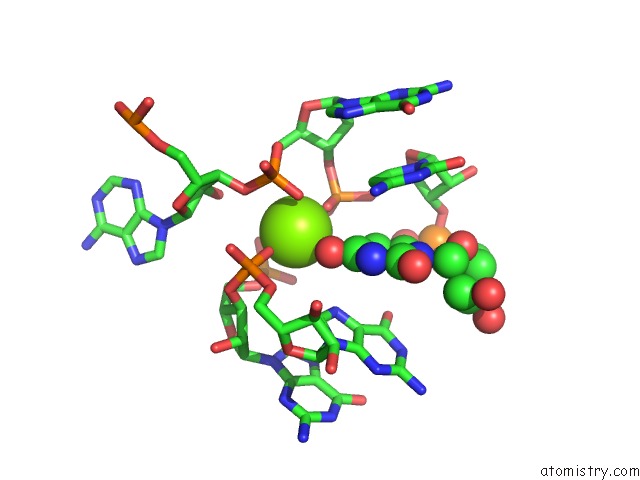
Mono view
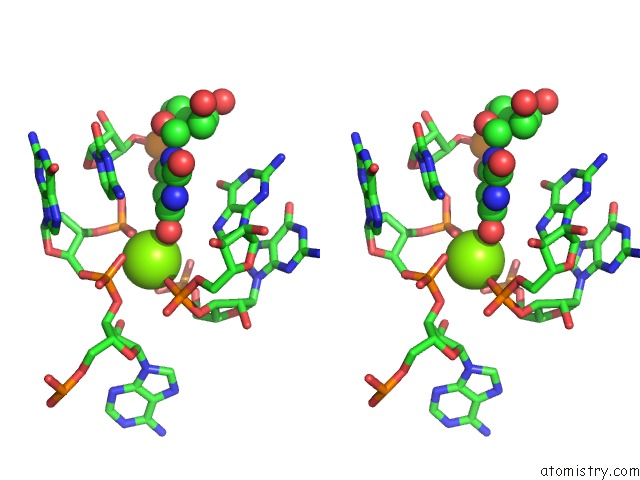
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of Human Pir-Mirna-300 Apical Loop Fused to the Ydao Riboswitch Scaffold within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 6wtr
Go back to
Magnesium binding site 2 out
of 3 in the Structure of Human Pir-Mirna-300 Apical Loop Fused to the Ydao Riboswitch Scaffold
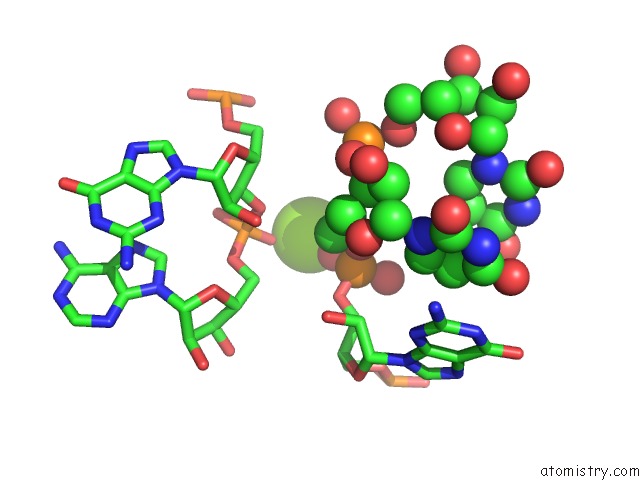
Mono view
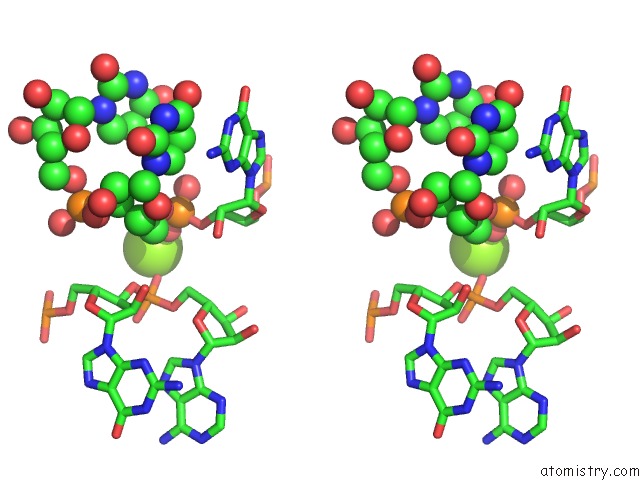
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of Human Pir-Mirna-300 Apical Loop Fused to the Ydao Riboswitch Scaffold within 5.0Å range:
|
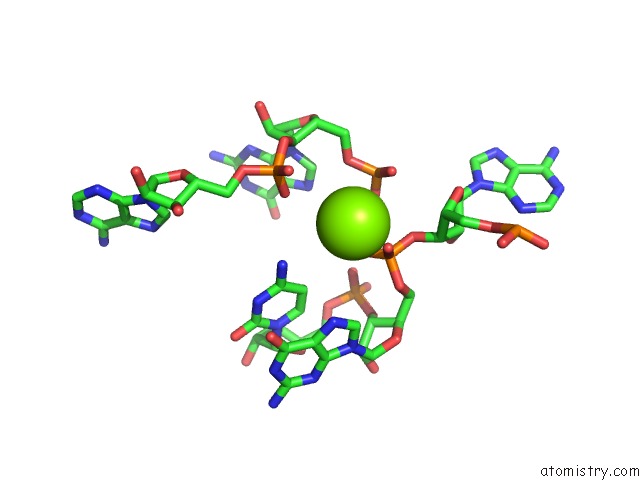
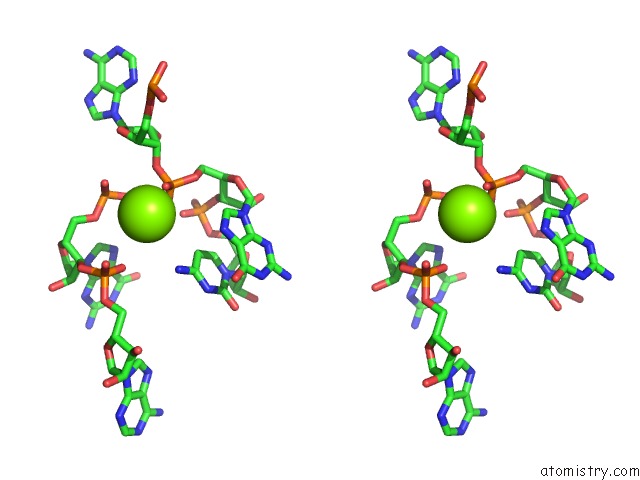
Magnesium binding site 3 out of 3 in 6wtr
Go back to
Magnesium binding site 3 out
of 3 in the Structure of Human Pir-Mirna-300 Apical Loop Fused to the Ydao Riboswitch Scaffold
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structure of Human Pir-Mirna-300 Apical Loop Fused to the Ydao Riboswitch Scaffold within 5.0Å range:
|
Reference:
G.M.Shoffner,
Z.Peng,
F.Guo.
Structures of Microrna-Precursor Apical Junctions and Loops Revealed By Scaffold-Directed Crystallography: Importance of Non-Canonical Base Pairs in Processing To Be Published.
Page generated: Tue Oct 1 23:14:06 2024
Last articles
Cl in 8AKCCl in 8AK9
Cl in 8AK8
Cl in 8AK7
Cl in 8AJP
Cl in 8AJ6
Cl in 8AIO
Cl in 8AI7
Cl in 8AHZ
Cl in 8AHQ