Magnesium »
PDB 6xmt-6y0t »
6xny »
Magnesium in PDB 6xny: Structure of RAG1 (R848M/E649V)-RAG2-Dna Strand Transfer Complex (Paired-Form)
Enzymatic activity of Structure of RAG1 (R848M/E649V)-RAG2-Dna Strand Transfer Complex (Paired-Form)
All present enzymatic activity of Structure of RAG1 (R848M/E649V)-RAG2-Dna Strand Transfer Complex (Paired-Form):
2.3.2.27;
2.3.2.27;
Other elements in 6xny:
The structure of Structure of RAG1 (R848M/E649V)-RAG2-Dna Strand Transfer Complex (Paired-Form) also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of RAG1 (R848M/E649V)-RAG2-Dna Strand Transfer Complex (Paired-Form)
(pdb code 6xny). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Structure of RAG1 (R848M/E649V)-RAG2-Dna Strand Transfer Complex (Paired-Form), PDB code: 6xny:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Structure of RAG1 (R848M/E649V)-RAG2-Dna Strand Transfer Complex (Paired-Form), PDB code: 6xny:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 6xny
Go back to
Magnesium binding site 1 out
of 4 in the Structure of RAG1 (R848M/E649V)-RAG2-Dna Strand Transfer Complex (Paired-Form)
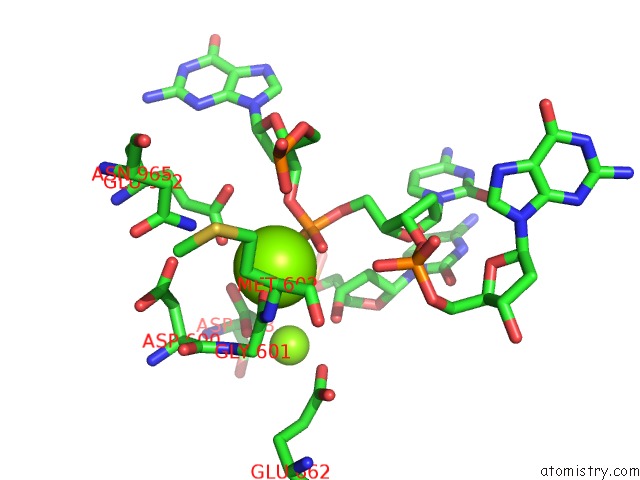
Mono view
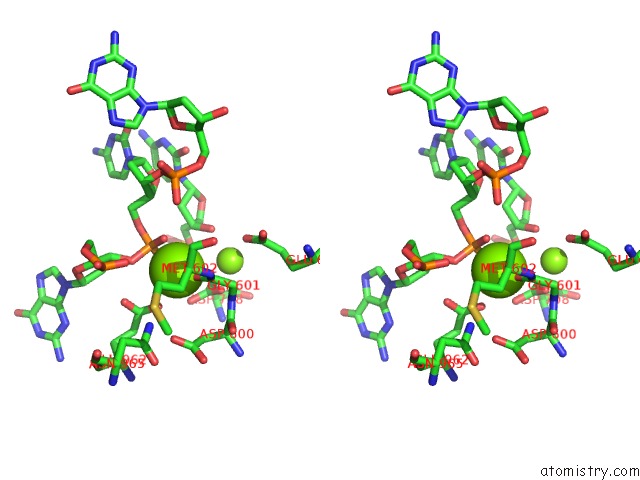
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of RAG1 (R848M/E649V)-RAG2-Dna Strand Transfer Complex (Paired-Form) within 5.0Å range:
|
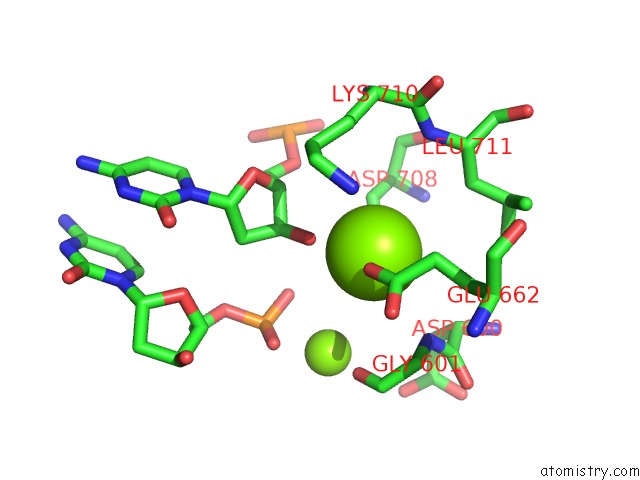
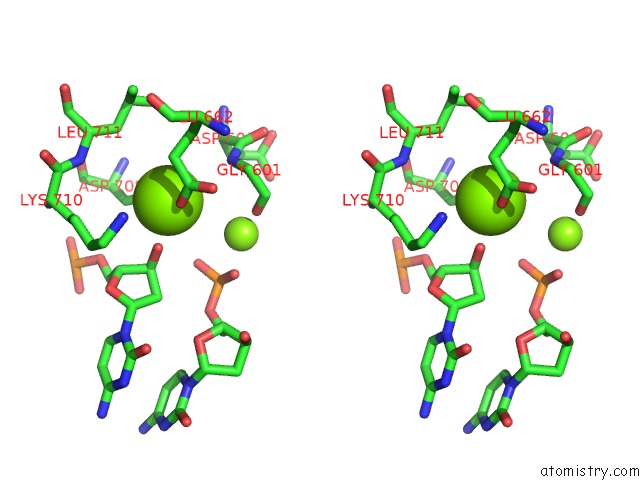
Magnesium binding site 2 out of 4 in 6xny
Go back to
Magnesium binding site 2 out
of 4 in the Structure of RAG1 (R848M/E649V)-RAG2-Dna Strand Transfer Complex (Paired-Form)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of RAG1 (R848M/E649V)-RAG2-Dna Strand Transfer Complex (Paired-Form) within 5.0Å range:
|
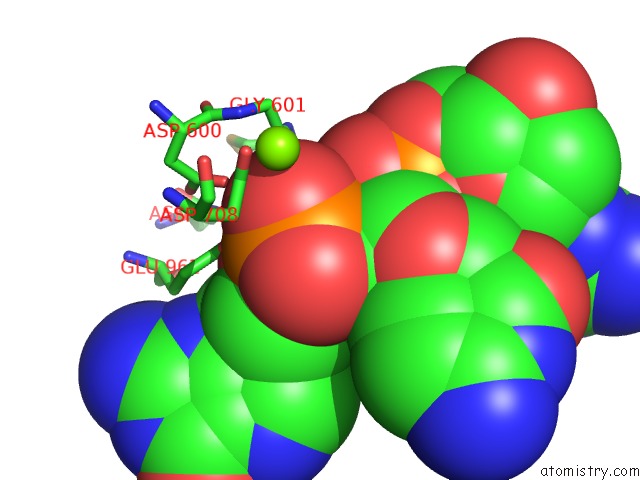
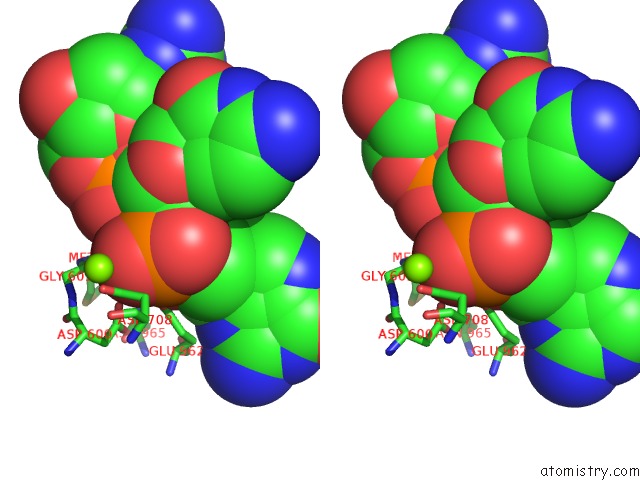
Magnesium binding site 3 out of 4 in 6xny
Go back to
Magnesium binding site 3 out
of 4 in the Structure of RAG1 (R848M/E649V)-RAG2-Dna Strand Transfer Complex (Paired-Form)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structure of RAG1 (R848M/E649V)-RAG2-Dna Strand Transfer Complex (Paired-Form) within 5.0Å range:
|
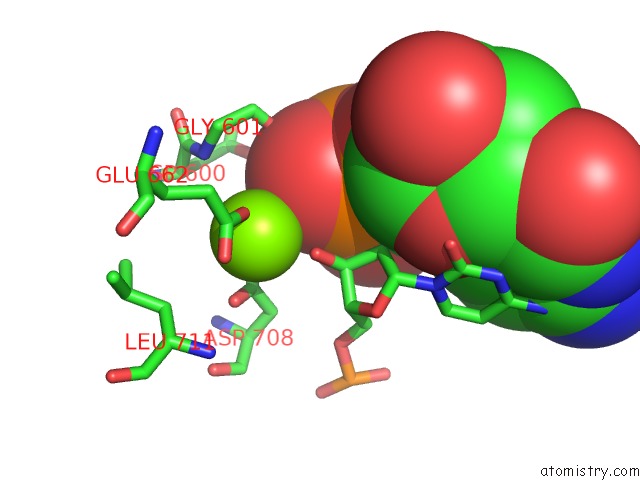
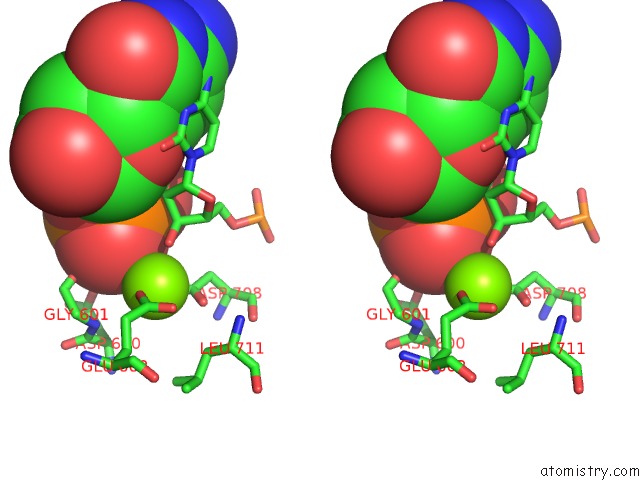
Magnesium binding site 4 out of 4 in 6xny
Go back to
Magnesium binding site 4 out
of 4 in the Structure of RAG1 (R848M/E649V)-RAG2-Dna Strand Transfer Complex (Paired-Form)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Structure of RAG1 (R848M/E649V)-RAG2-Dna Strand Transfer Complex (Paired-Form) within 5.0Å range:
|
Reference:
Y.Zhang,
E.Corbett,
S.Wu,
D.G.Schatz.
Structural Basis For the Activation and Suppression of Transposition During Evolution of the Rag Recombinase. Embo J. V. 39 05857 2020.
ISSN: ESSN 1460-2075
PubMed: 32945578
DOI: 10.15252/EMBJ.2020105857
Page generated: Tue Oct 1 23:37:04 2024
ISSN: ESSN 1460-2075
PubMed: 32945578
DOI: 10.15252/EMBJ.2020105857
Last articles
Cl in 3SLFCl in 3SKH
Cl in 3SIY
Cl in 3SJX
Cl in 3SK0
Cl in 3SHX
Cl in 3SJG
Cl in 3SJF
Cl in 3SJE
Cl in 3SIX