Magnesium »
PDB 6xmt-6y0t »
6xrc »
Magnesium in PDB 6xrc: Apo Nis Synthetase Desd Variant R306Q
Protein crystallography data
The structure of Apo Nis Synthetase Desd Variant R306Q, PDB code: 6xrc
was solved by
K.M.Hoffmann,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 59.47 / 2.45 |
| Space group | P 2 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.417, 98.921, 183.352, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20 / 27 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Apo Nis Synthetase Desd Variant R306Q
(pdb code 6xrc). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Apo Nis Synthetase Desd Variant R306Q, PDB code: 6xrc:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Apo Nis Synthetase Desd Variant R306Q, PDB code: 6xrc:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 6xrc
Go back to
Magnesium binding site 1 out
of 3 in the Apo Nis Synthetase Desd Variant R306Q
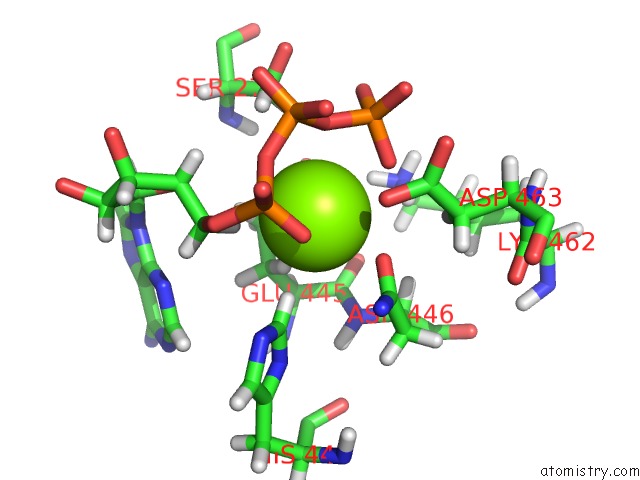
Mono view
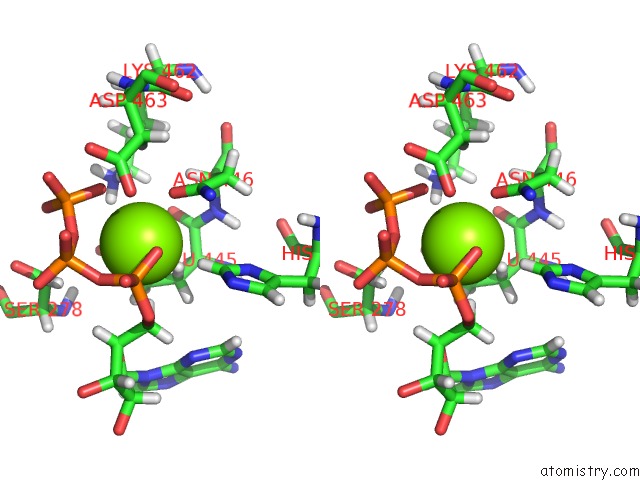
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Apo Nis Synthetase Desd Variant R306Q within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 6xrc
Go back to
Magnesium binding site 2 out
of 3 in the Apo Nis Synthetase Desd Variant R306Q
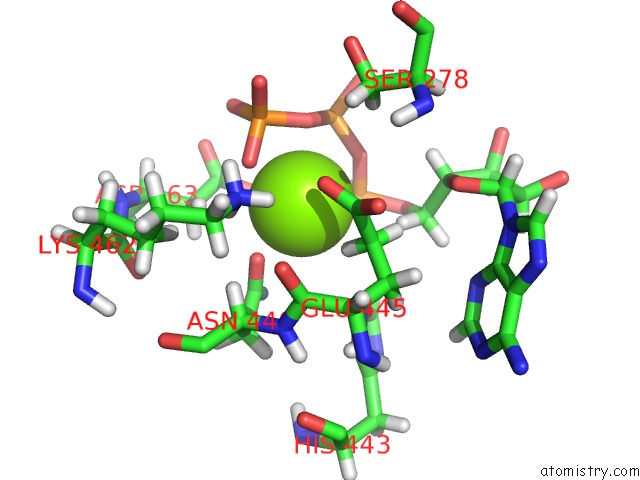
Mono view
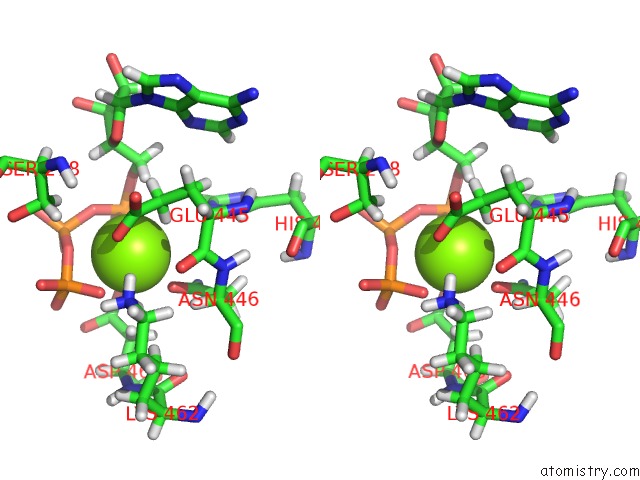
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Apo Nis Synthetase Desd Variant R306Q within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 6xrc
Go back to
Magnesium binding site 3 out
of 3 in the Apo Nis Synthetase Desd Variant R306Q
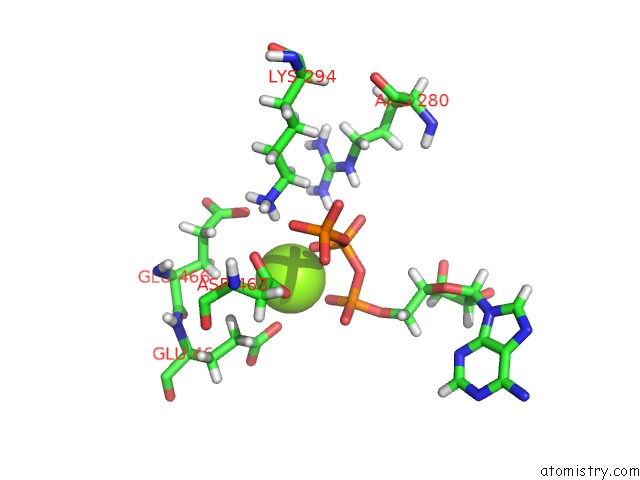
Mono view
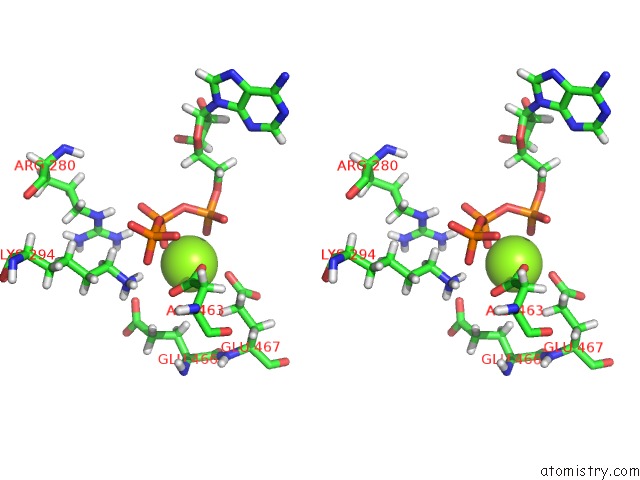
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Apo Nis Synthetase Desd Variant R306Q within 5.0Å range:
|
Reference:
K.M.Hoffmann,
E.S.Goncuian,
K.L.Karimi,
C.R.Amendola,
Y.Mojab,
K.M.Wood,
G.A.Prussia,
J.Nix,
M.Yamamoto,
K.Lathan,
I.W.Orion.
Cofactor Complexes of Desd, A Model Enzyme in the Virulence-Related Nis Synthetase Family. Biochemistry V. 59 3427 2020.
ISSN: ISSN 0006-2960
PubMed: 32885650
DOI: 10.1021/ACS.BIOCHEM.9B00899
Page generated: Tue Oct 1 23:38:46 2024
ISSN: ISSN 0006-2960
PubMed: 32885650
DOI: 10.1021/ACS.BIOCHEM.9B00899
Last articles
Cl in 3S00Cl in 3RZ3
Cl in 3RXY
Cl in 3RZF
Cl in 3RW8
Cl in 3RXZ
Cl in 3RVB
Cl in 3RV4
Cl in 3RVF
Cl in 3RUL