Magnesium »
PDB 7a1x-7adt »
7aav »
Magnesium in PDB 7aav: Human Pre-Bact-2 Spliceosome Core Structure
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Human Pre-Bact-2 Spliceosome Core Structure
(pdb code 7aav). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Human Pre-Bact-2 Spliceosome Core Structure, PDB code: 7aav:
In total only one binding site of Magnesium was determined in the Human Pre-Bact-2 Spliceosome Core Structure, PDB code: 7aav:
Magnesium binding site 1 out of 1 in 7aav
Go back to
Magnesium binding site 1 out
of 1 in the Human Pre-Bact-2 Spliceosome Core Structure
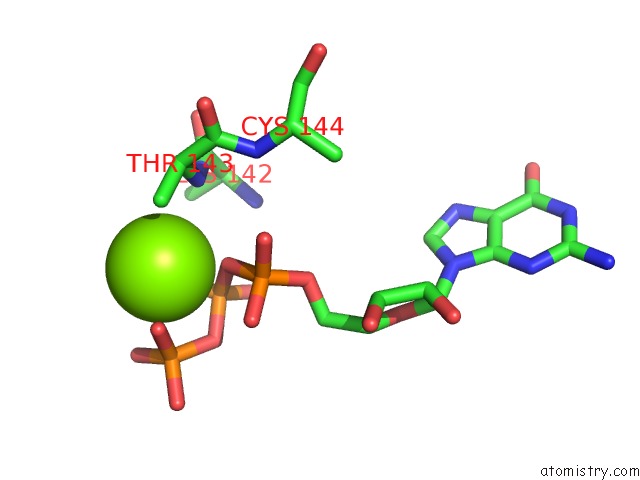
Mono view
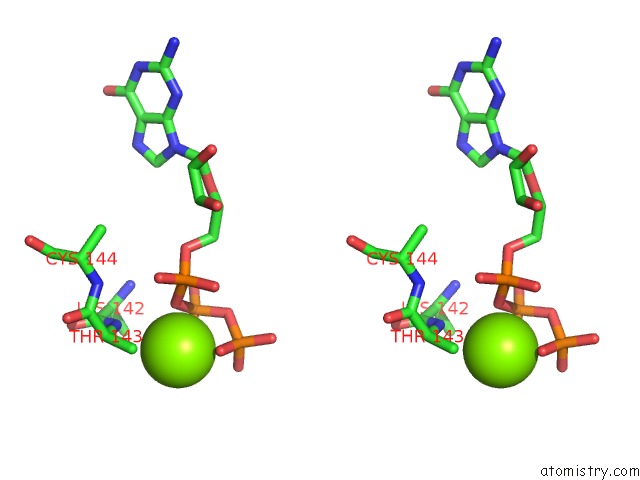
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Human Pre-Bact-2 Spliceosome Core Structure within 5.0Å range:
|
Reference:
C.Townsend,
M.N.Leelaram,
D.E.Agafonov,
O.Dybkov,
C.L.Will,
K.Bertram,
H.Urlaub,
B.Kastner,
H.Stark,
R.Luhrmann.
Mechanism of Protein-Guided Folding of the Active Site U2/U6 Rna During Spliceosome Activation. Science 2020.
ISSN: ESSN 1095-9203
PubMed: 33243851
DOI: 10.1126/SCIENCE.ABC3753
Page generated: Wed Oct 2 09:08:16 2024
ISSN: ESSN 1095-9203
PubMed: 33243851
DOI: 10.1126/SCIENCE.ABC3753
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1