Magnesium »
PDB 7diy-7dui »
7dph »
Magnesium in PDB 7dph: H-Ras Q61H in Complex with Gppnhp (State 1) After Structural Transition By Humidity Control
Enzymatic activity of H-Ras Q61H in Complex with Gppnhp (State 1) After Structural Transition By Humidity Control
All present enzymatic activity of H-Ras Q61H in Complex with Gppnhp (State 1) After Structural Transition By Humidity Control:
3.6.5.2;
3.6.5.2;
Protein crystallography data
The structure of H-Ras Q61H in Complex with Gppnhp (State 1) After Structural Transition By Humidity Control, PDB code: 7dph
was solved by
H.Taniguchi,
S.Matsumoto,
T.Kawamura,
T.Kumasaka,
T.Kataoka,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 23.21 / 1.54 |
| Space group | H 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 91.769, 91.769, 121.268, 90, 90, 120 |
| R / Rfree (%) | 19.2 / 20.2 |
Other elements in 7dph:
The structure of H-Ras Q61H in Complex with Gppnhp (State 1) After Structural Transition By Humidity Control also contains other interesting chemical elements:
| Sodium | (Na) | 1 atom |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the H-Ras Q61H in Complex with Gppnhp (State 1) After Structural Transition By Humidity Control
(pdb code 7dph). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the H-Ras Q61H in Complex with Gppnhp (State 1) After Structural Transition By Humidity Control, PDB code: 7dph:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the H-Ras Q61H in Complex with Gppnhp (State 1) After Structural Transition By Humidity Control, PDB code: 7dph:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 7dph
Go back to
Magnesium binding site 1 out
of 2 in the H-Ras Q61H in Complex with Gppnhp (State 1) After Structural Transition By Humidity Control
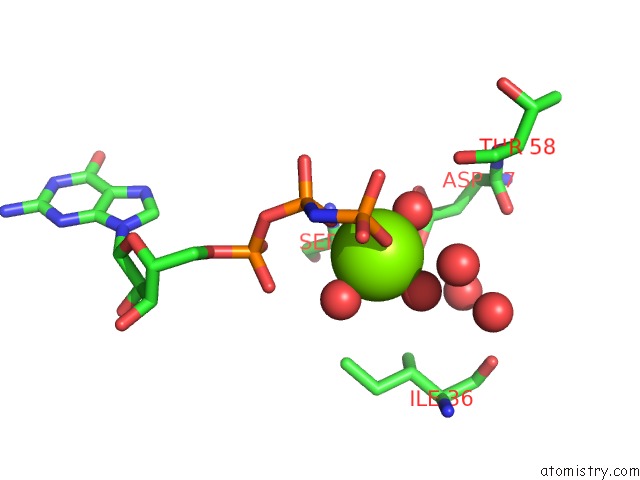
Mono view
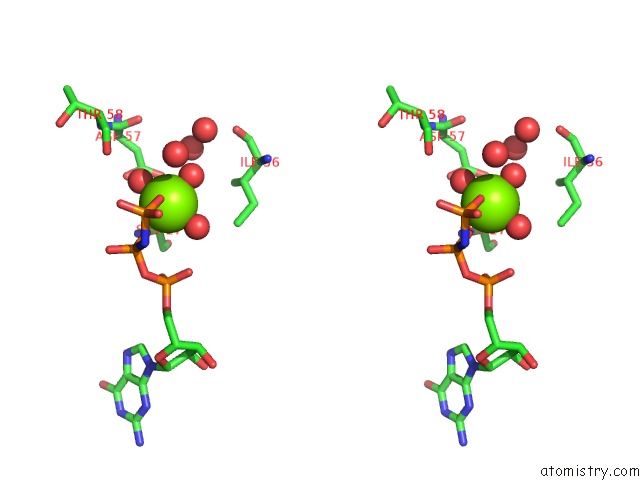
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of H-Ras Q61H in Complex with Gppnhp (State 1) After Structural Transition By Humidity Control within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 7dph
Go back to
Magnesium binding site 2 out
of 2 in the H-Ras Q61H in Complex with Gppnhp (State 1) After Structural Transition By Humidity Control
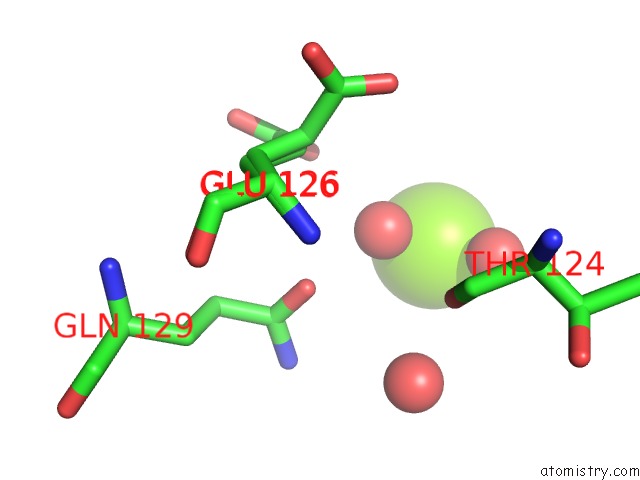
Mono view
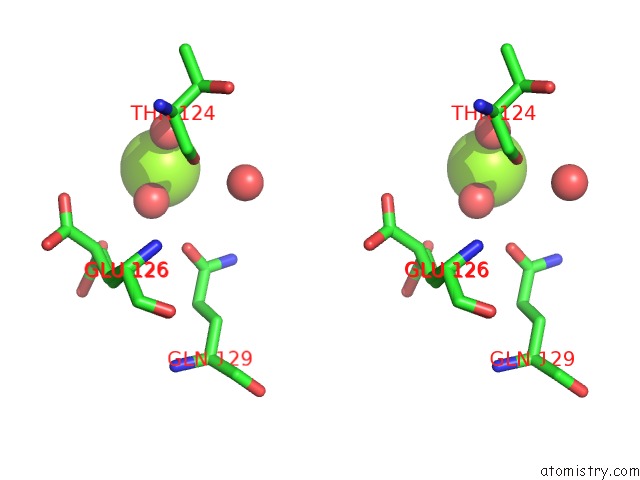
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of H-Ras Q61H in Complex with Gppnhp (State 1) After Structural Transition By Humidity Control within 5.0Å range:
|
Reference:
S.Matsumoto,
H.Taniguchi-Tamura,
M.Araki,
T.Kawamura,
R.Miyamoto,
C.Tsuda,
F.Shima,
T.Kumasaka,
Y.Okuno,
T.Kataoka.
Oncogenic Mutations Q61L and Q61H Confer Active Form-Like Structural Features to the Inactive State (State 1) Conformation of H-Ras Protein. Biochem.Biophys.Res.Commun. V. 565 85 2021.
ISSN: ESSN 1090-2104
PubMed: 34102474
DOI: 10.1016/J.BBRC.2021.05.084
Page generated: Wed Oct 2 15:57:51 2024
ISSN: ESSN 1090-2104
PubMed: 34102474
DOI: 10.1016/J.BBRC.2021.05.084
Last articles
Ca in 5OWOCa in 5OWR
Ca in 5OWC
Ca in 5OW8
Ca in 5OTJ
Ca in 5OSQ
Ca in 5OTN
Ca in 5OV7
Ca in 5OT1
Ca in 5ONR