Magnesium »
PDB 7e21-7eg1 »
7e51 »
Magnesium in PDB 7e51: Structure of Pep Bound Enolase From Mycobacterium Tuberculosis
Enzymatic activity of Structure of Pep Bound Enolase From Mycobacterium Tuberculosis
All present enzymatic activity of Structure of Pep Bound Enolase From Mycobacterium Tuberculosis:
4.2.1.11;
4.2.1.11;
Magnesium Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 16;Binding sites:
The binding sites of Magnesium atom in the Structure of Pep Bound Enolase From Mycobacterium Tuberculosis (pdb code 7e51). This binding sites where shown within 5.0 Angstroms radius around Magnesium atom.In total 16 binding sites of Magnesium where determined in the Structure of Pep Bound Enolase From Mycobacterium Tuberculosis, PDB code: 7e51:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
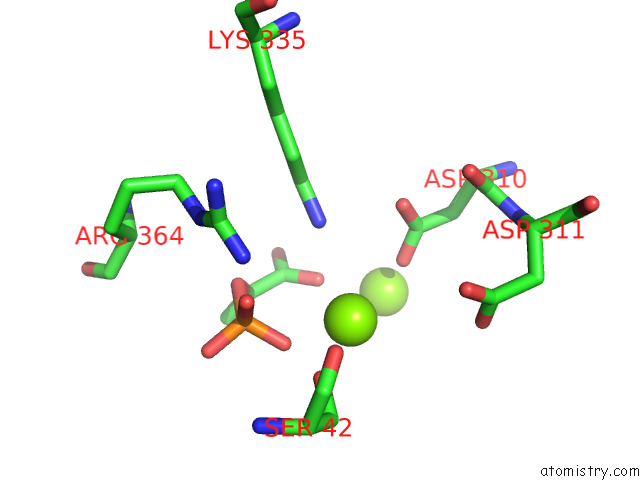
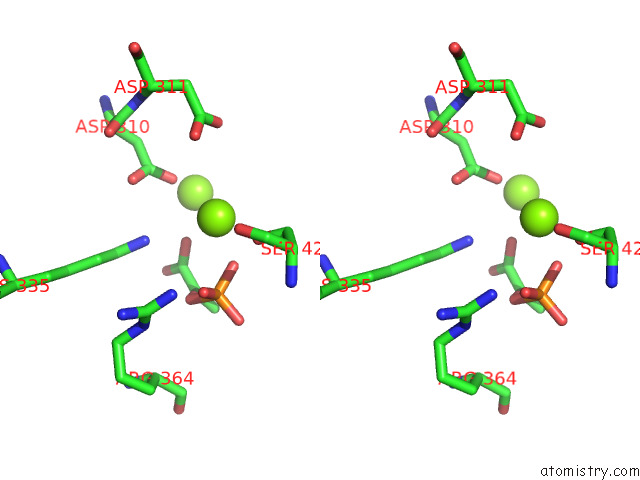
Magnesium binding site 1 out of 16 in 7e51
Go back to
Magnesium binding site 1 out
of 16 in the Structure of Pep Bound Enolase From Mycobacterium Tuberculosis
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of Pep Bound Enolase From Mycobacterium Tuberculosis within 5.0Å range:
|
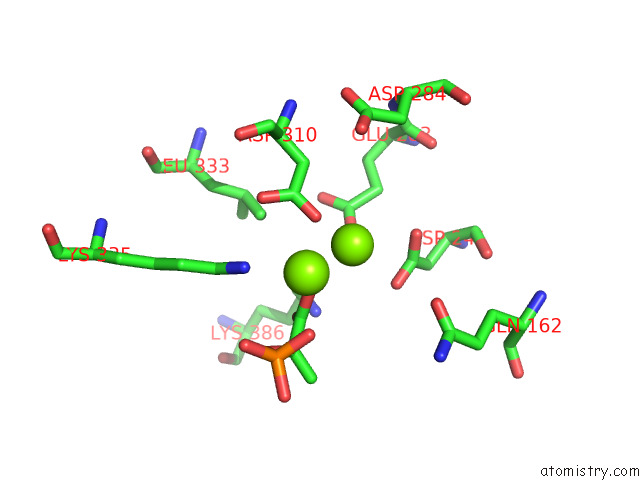
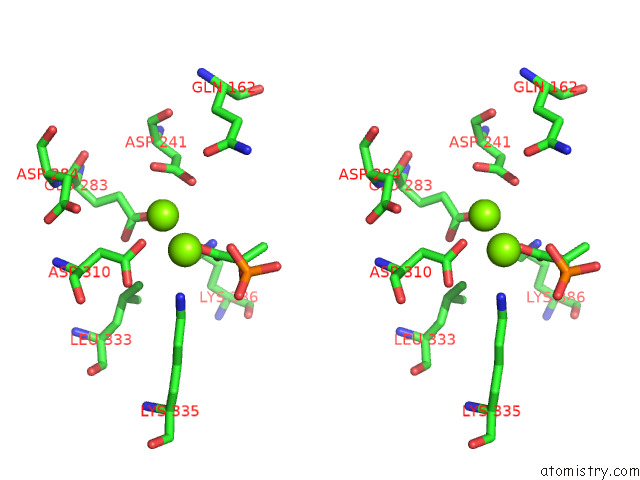
Magnesium binding site 2 out of 16 in 7e51
Go back to
Magnesium binding site 2 out
of 16 in the Structure of Pep Bound Enolase From Mycobacterium Tuberculosis
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of Pep Bound Enolase From Mycobacterium Tuberculosis within 5.0Å range:
|
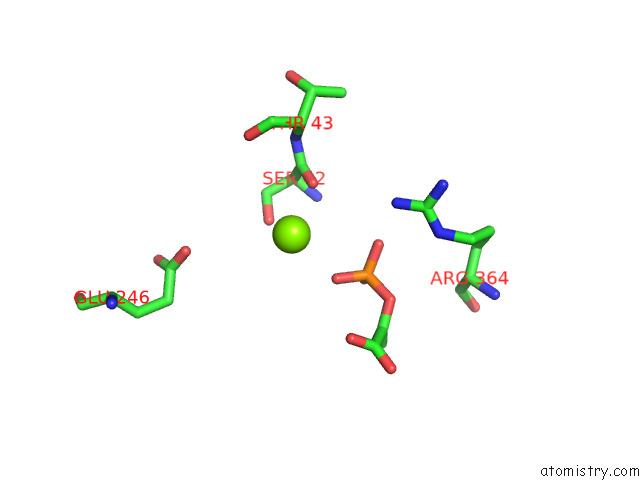
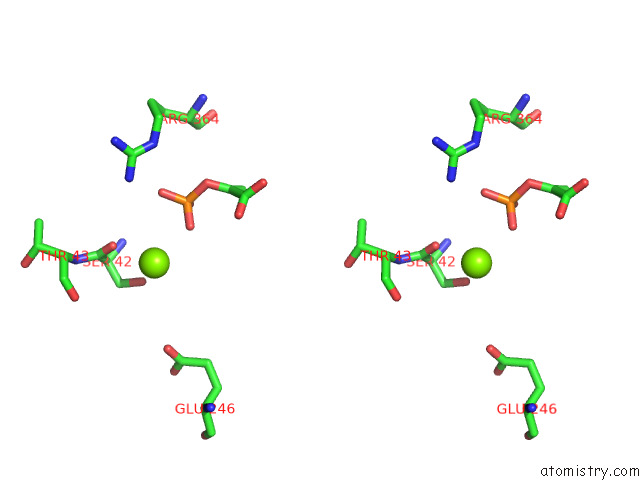
Magnesium binding site 3 out of 16 in 7e51
Go back to
Magnesium binding site 3 out
of 16 in the Structure of Pep Bound Enolase From Mycobacterium Tuberculosis
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structure of Pep Bound Enolase From Mycobacterium Tuberculosis within 5.0Å range:
|
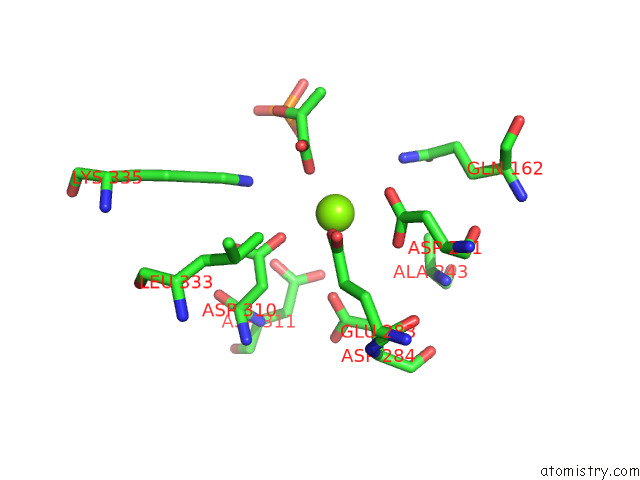
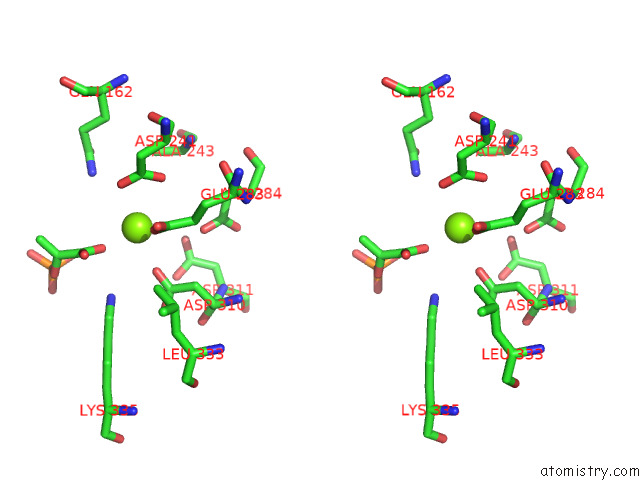
Magnesium binding site 4 out of 16 in 7e51
Go back to
Magnesium binding site 4 out
of 16 in the Structure of Pep Bound Enolase From Mycobacterium Tuberculosis
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Structure of Pep Bound Enolase From Mycobacterium Tuberculosis within 5.0Å range:
|
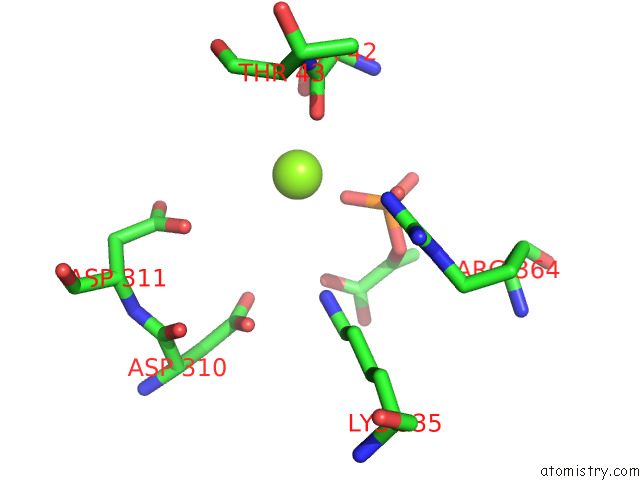
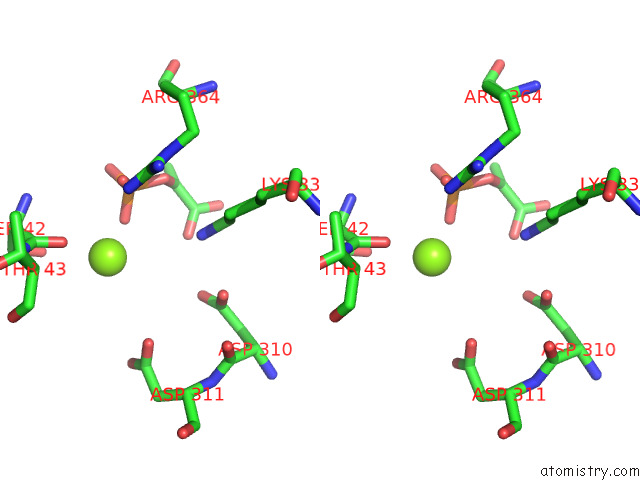
Magnesium binding site 5 out of 16 in 7e51
Go back to
Magnesium binding site 5 out
of 16 in the Structure of Pep Bound Enolase From Mycobacterium Tuberculosis
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Structure of Pep Bound Enolase From Mycobacterium Tuberculosis within 5.0Å range:
|
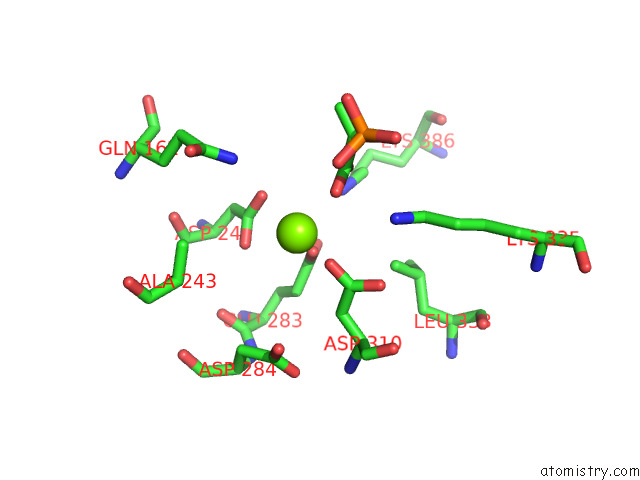
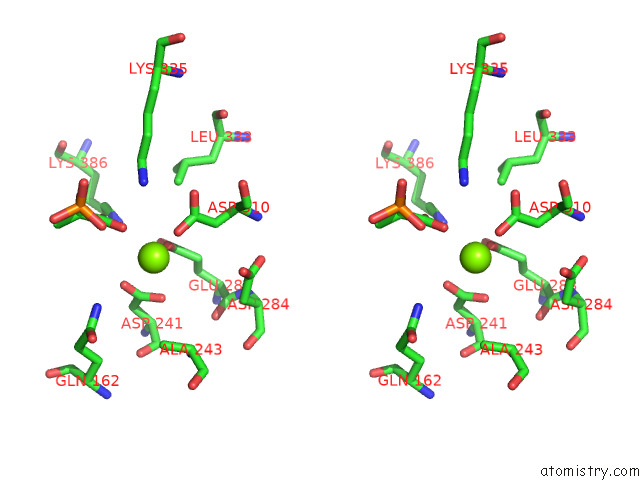
Magnesium binding site 6 out of 16 in 7e51
Go back to
Magnesium binding site 6 out
of 16 in the Structure of Pep Bound Enolase From Mycobacterium Tuberculosis
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 6 of Structure of Pep Bound Enolase From Mycobacterium Tuberculosis within 5.0Å range:
|
Magnesium binding site 7 out of 16 in 7e51
Go back to
Magnesium binding site 7 out
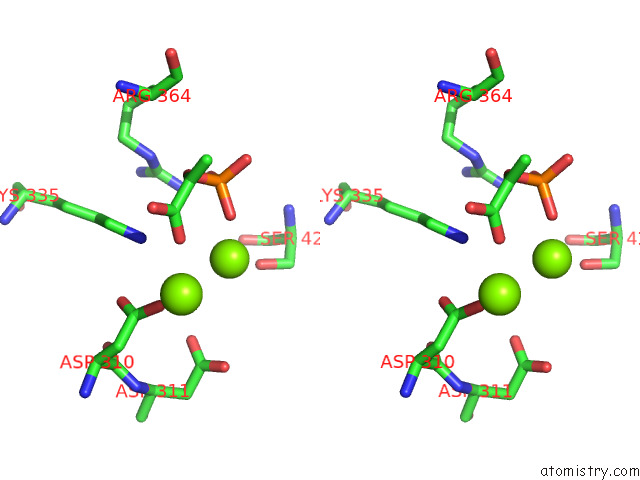
of 16 in the Structure of Pep Bound Enolase From Mycobacterium Tuberculosis
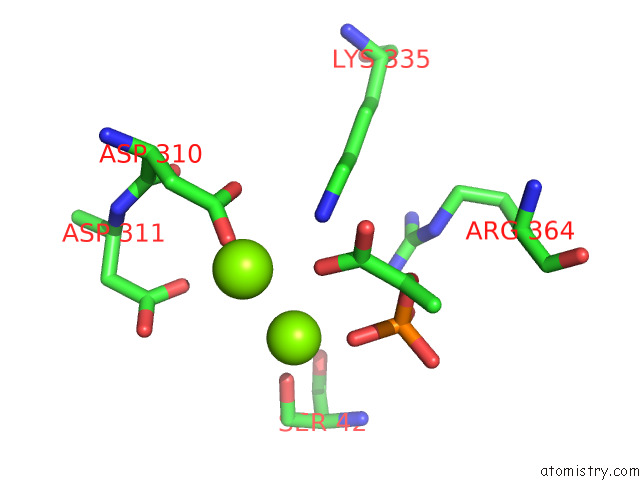
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 7 of Structure of Pep Bound Enolase From Mycobacterium Tuberculosis within 5.0Å range:
|
Magnesium binding site 8 out of 16 in 7e51
Go back to
Magnesium binding site 8 out
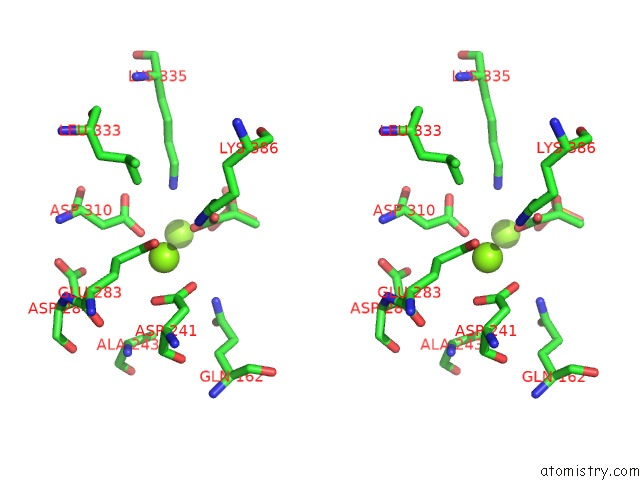
of 16 in the Structure of Pep Bound Enolase From Mycobacterium Tuberculosis
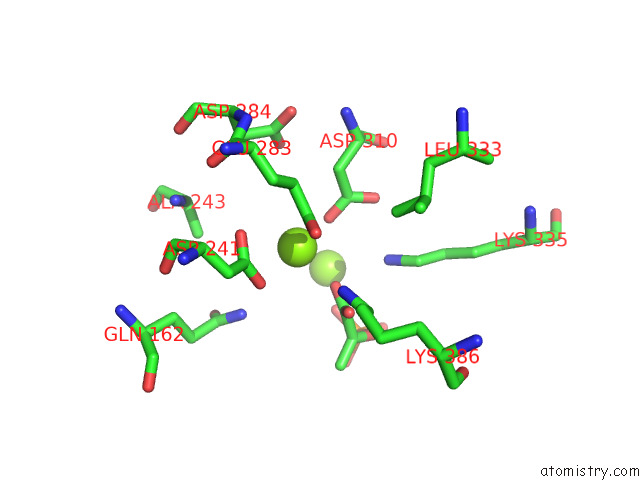
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 8 of Structure of Pep Bound Enolase From Mycobacterium Tuberculosis within 5.0Å range:
|
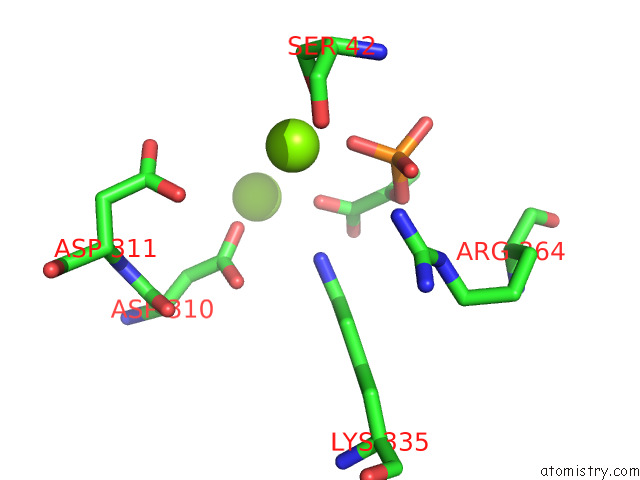
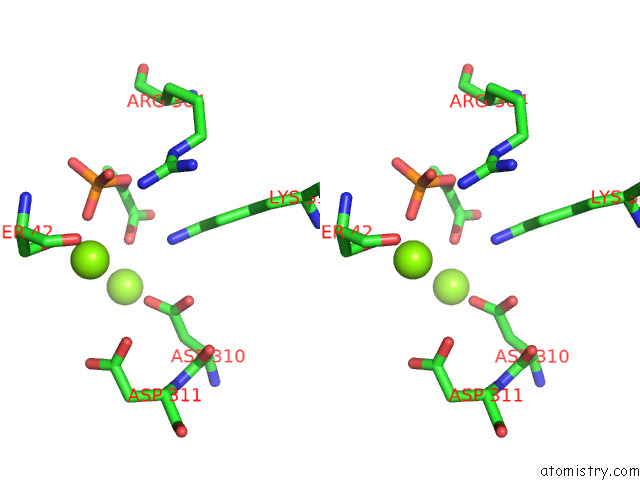
Magnesium binding site 9 out of 16 in 7e51
Go back to
Magnesium binding site 9 out
of 16 in the Structure of Pep Bound Enolase From Mycobacterium Tuberculosis
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 9 of Structure of Pep Bound Enolase From Mycobacterium Tuberculosis within 5.0Å range:
|
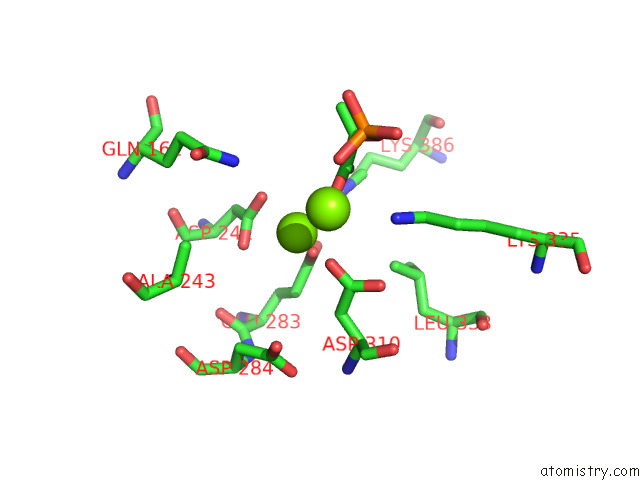
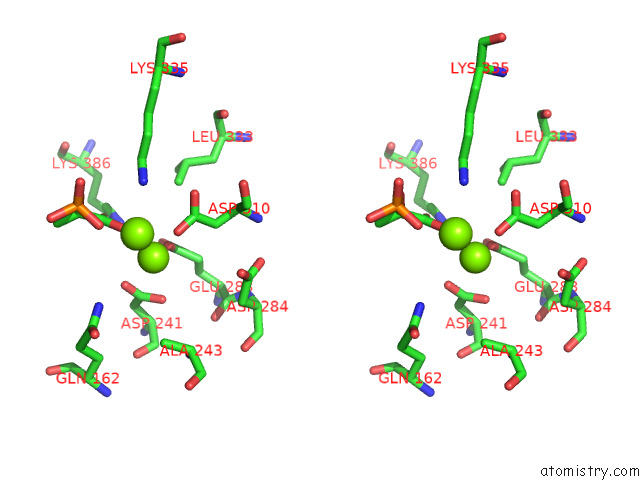
Magnesium binding site 10 out of 16 in 7e51
Go back to
Magnesium binding site 10 out
of 16 in the Structure of Pep Bound Enolase From Mycobacterium Tuberculosis
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 10 of Structure of Pep Bound Enolase From Mycobacterium Tuberculosis within 5.0Å range:
|
Reference:
M.Ahmad,
B.Jha,
S.Bose,
R.Mariadasee,
A.Dwivedy,
S.Tiwari,
R.Pal,
D.Kar,
T.Parish,
J.Jeyakanthan,
K.R.Vinothkumar,
B.K.Biswal.
Structural Snapshots of Mycobacterium Tuberculosis Enolase Reveal Dual Mode of 2PG Binding and Its Implication in Enzyme Catalysis. To Be Published.
Page generated: Wed Oct 2 20:31:39 2024
Last articles
F in 7K77F in 7K8G
F in 7K89
F in 7K87
F in 7K6Z
F in 7K6M
F in 7K6A
F in 7K4D
F in 7K6L
F in 7K4F