Magnesium »
PDB 7e21-7eg1 »
7e9e »
Magnesium in PDB 7e9e: Crystal Structure of A Class I PREQ1 Riboswitch Aptamer (AB13-14) Complexed with A Cognate Ligand-Derived Photoaffinity Probe
Protein crystallography data
The structure of Crystal Structure of A Class I PREQ1 Riboswitch Aptamer (AB13-14) Complexed with A Cognate Ligand-Derived Photoaffinity Probe, PDB code: 7e9e
was solved by
T.Numata,
J.S.Schneekloth,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.60 / 1.57 |
| Space group | P 61 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 52.691, 52.691, 176.999, 90, 90, 120 |
| R / Rfree (%) | 19 / 20.8 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of A Class I PREQ1 Riboswitch Aptamer (AB13-14) Complexed with A Cognate Ligand-Derived Photoaffinity Probe
(pdb code 7e9e). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Crystal Structure of A Class I PREQ1 Riboswitch Aptamer (AB13-14) Complexed with A Cognate Ligand-Derived Photoaffinity Probe, PDB code: 7e9e:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Crystal Structure of A Class I PREQ1 Riboswitch Aptamer (AB13-14) Complexed with A Cognate Ligand-Derived Photoaffinity Probe, PDB code: 7e9e:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 7e9e
Go back to
Magnesium binding site 1 out
of 3 in the Crystal Structure of A Class I PREQ1 Riboswitch Aptamer (AB13-14) Complexed with A Cognate Ligand-Derived Photoaffinity Probe
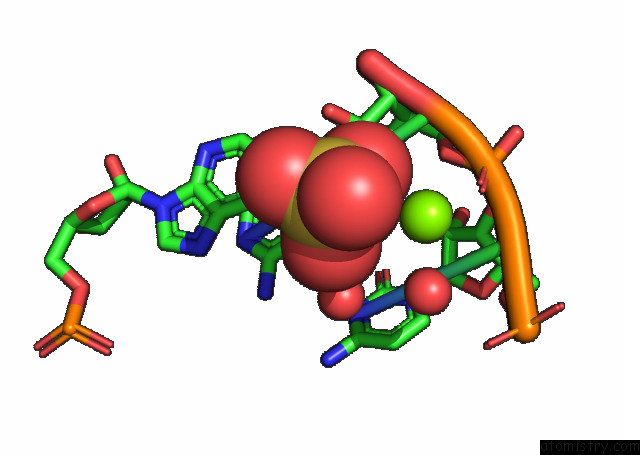
Mono view
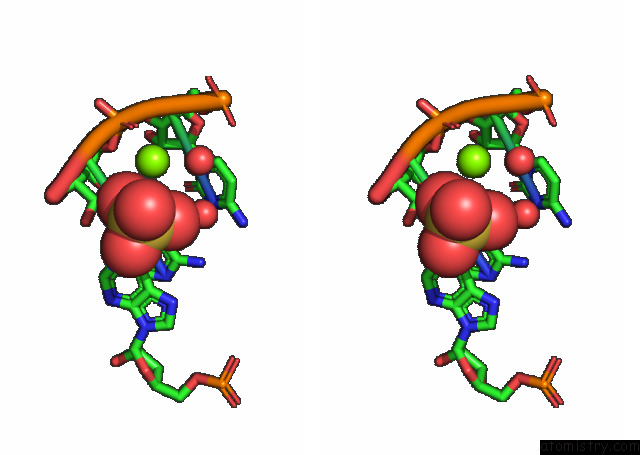
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of A Class I PREQ1 Riboswitch Aptamer (AB13-14) Complexed with A Cognate Ligand-Derived Photoaffinity Probe within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 7e9e
Go back to
Magnesium binding site 2 out
of 3 in the Crystal Structure of A Class I PREQ1 Riboswitch Aptamer (AB13-14) Complexed with A Cognate Ligand-Derived Photoaffinity Probe
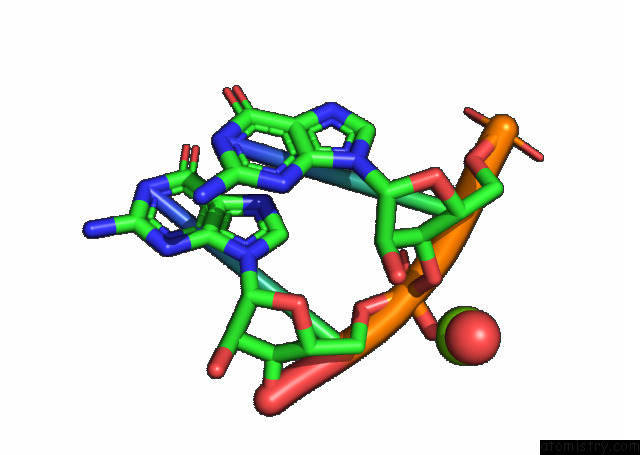
Mono view
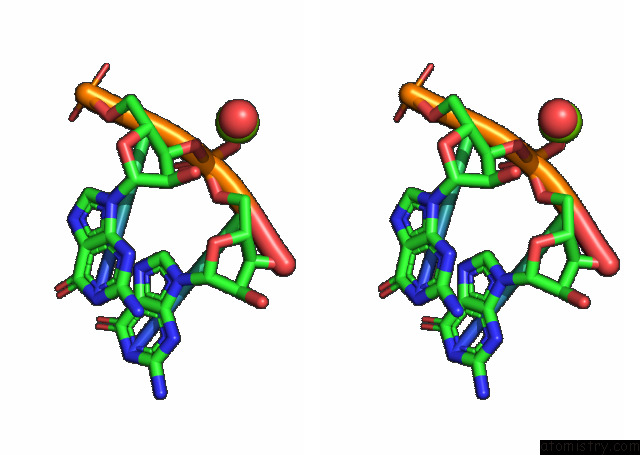
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of A Class I PREQ1 Riboswitch Aptamer (AB13-14) Complexed with A Cognate Ligand-Derived Photoaffinity Probe within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 7e9e
Go back to
Magnesium binding site 3 out
of 3 in the Crystal Structure of A Class I PREQ1 Riboswitch Aptamer (AB13-14) Complexed with A Cognate Ligand-Derived Photoaffinity Probe
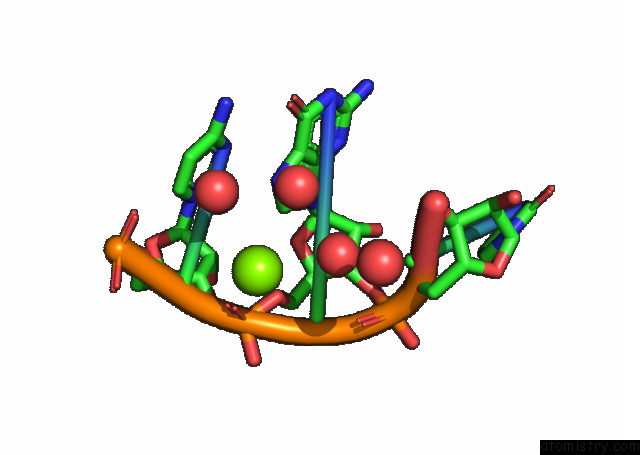
Mono view
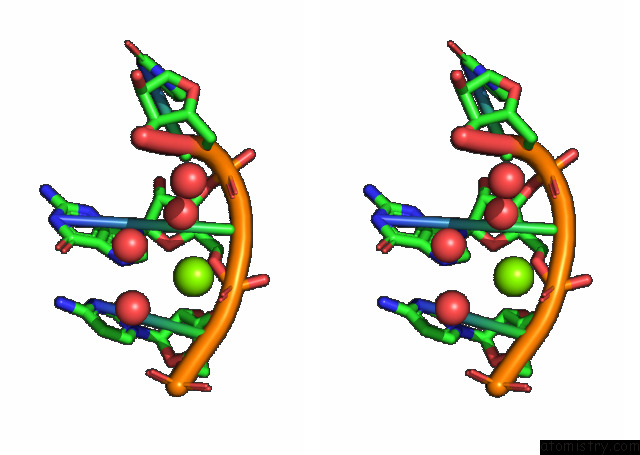
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of A Class I PREQ1 Riboswitch Aptamer (AB13-14) Complexed with A Cognate Ligand-Derived Photoaffinity Probe within 5.0Å range:
|
Reference:
S.Balaratnam,
C.Rhodes,
D.D.Bume,
C.Connelly,
C.C.Lai,
J.A.Kelley,
K.Yazdani,
P.J.Homan,
D.Incarnato,
T.Numata,
J.S.Schneekloth Jr..
A Chemical Probe Based on the Preq 1 Metabolite Enables Transcriptome-Wide Mapping of Binding Sites. Nat Commun V. 12 5856 2021.
ISSN: ESSN 2041-1723
PubMed: 34615874
DOI: 10.1038/S41467-021-25973-X
Page generated: Wed Oct 2 20:33:31 2024
ISSN: ESSN 2041-1723
PubMed: 34615874
DOI: 10.1038/S41467-021-25973-X
Last articles
Cl in 7U0ACl in 7TZX
Cl in 7TZW
Cl in 7TZY
Cl in 7TXR
Cl in 7TZ6
Cl in 7TZN
Cl in 7TZM
Cl in 7TXN
Cl in 7TXO