Magnesium »
PDB 7e20-7eg0 »
7ebc »
Magnesium in PDB 7ebc: Crystal Structure of Isocitrate Lyase-1 From Saccaromyces Cervisiae
Enzymatic activity of Crystal Structure of Isocitrate Lyase-1 From Saccaromyces Cervisiae
All present enzymatic activity of Crystal Structure of Isocitrate Lyase-1 From Saccaromyces Cervisiae:
4.1.3.1;
4.1.3.1;
Protein crystallography data
The structure of Crystal Structure of Isocitrate Lyase-1 From Saccaromyces Cervisiae, PDB code: 7ebc
was solved by
K.Hiragi,
K.Nishio,
S.Moriyama,
T.Hamaguchi,
A.Mizoguchi,
K.Yonekura,
K.Tani,
T.Mizushima,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.80 / 2.30 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 95.522, 129.377, 210.787, 90, 90, 90 |
| R / Rfree (%) | 18.9 / 22 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Isocitrate Lyase-1 From Saccaromyces Cervisiae
(pdb code 7ebc). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Crystal Structure of Isocitrate Lyase-1 From Saccaromyces Cervisiae, PDB code: 7ebc:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Crystal Structure of Isocitrate Lyase-1 From Saccaromyces Cervisiae, PDB code: 7ebc:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 7ebc
Go back to
Magnesium binding site 1 out
of 4 in the Crystal Structure of Isocitrate Lyase-1 From Saccaromyces Cervisiae
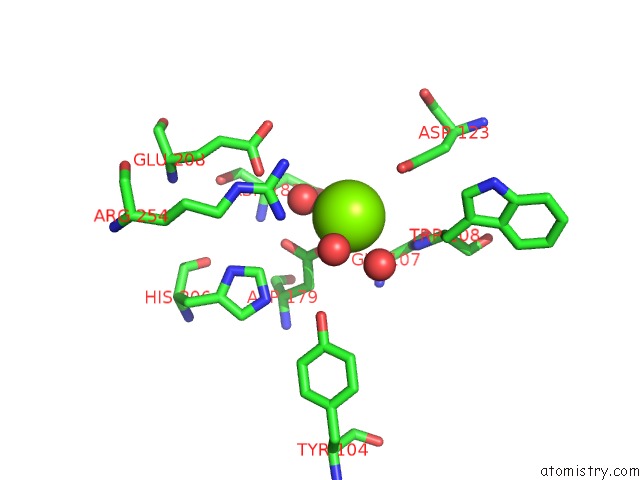
Mono view
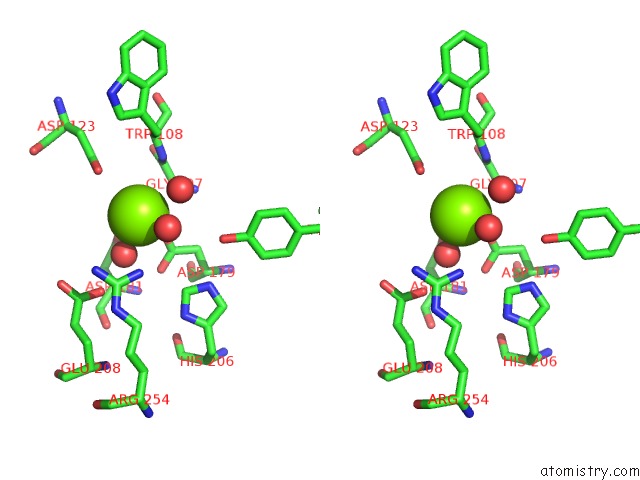
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Isocitrate Lyase-1 From Saccaromyces Cervisiae within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 7ebc
Go back to
Magnesium binding site 2 out
of 4 in the Crystal Structure of Isocitrate Lyase-1 From Saccaromyces Cervisiae
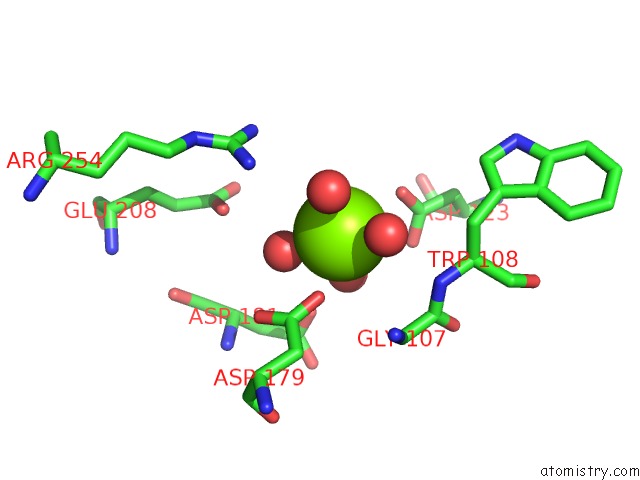
Mono view
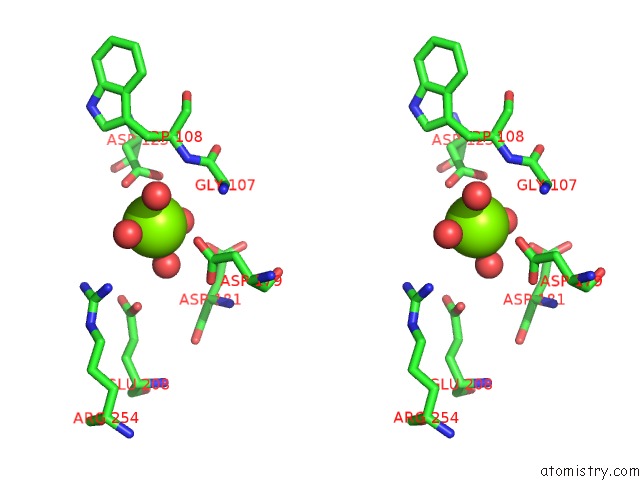
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Isocitrate Lyase-1 From Saccaromyces Cervisiae within 5.0Å range:
|
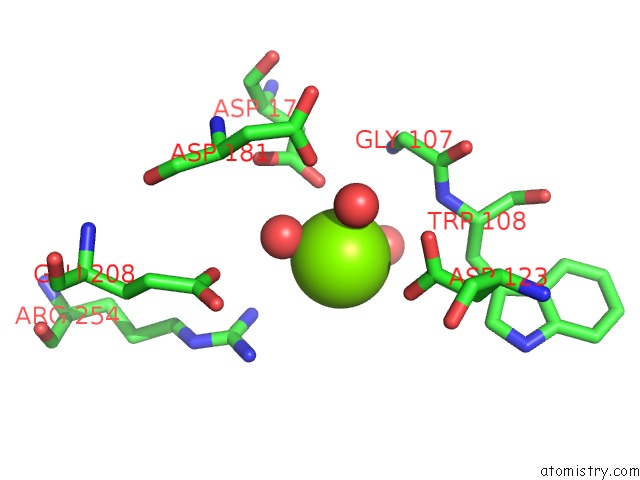
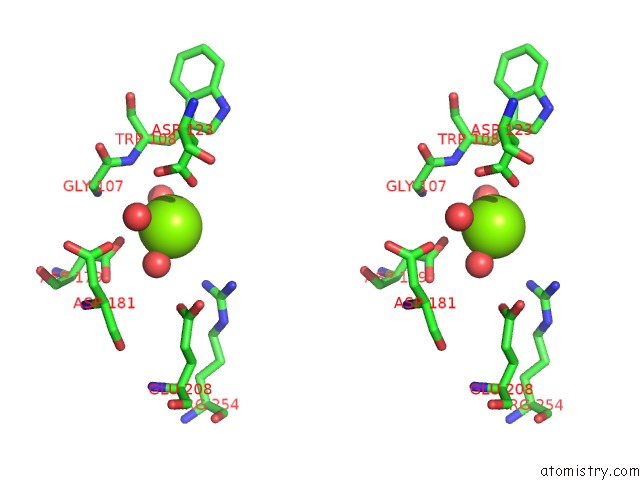
Magnesium binding site 3 out of 4 in 7ebc
Go back to
Magnesium binding site 3 out
of 4 in the Crystal Structure of Isocitrate Lyase-1 From Saccaromyces Cervisiae
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of Isocitrate Lyase-1 From Saccaromyces Cervisiae within 5.0Å range:
|
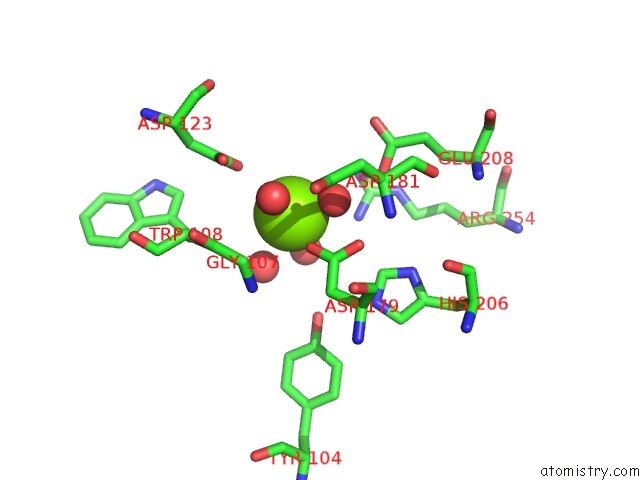
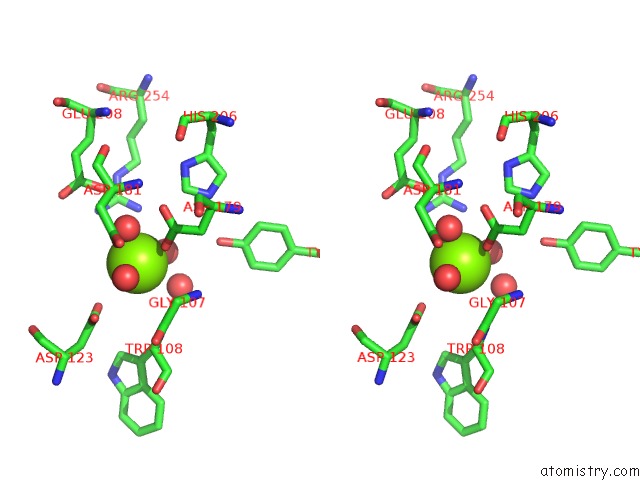
Magnesium binding site 4 out of 4 in 7ebc
Go back to
Magnesium binding site 4 out
of 4 in the Crystal Structure of Isocitrate Lyase-1 From Saccaromyces Cervisiae
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of Isocitrate Lyase-1 From Saccaromyces Cervisiae within 5.0Å range:
|
Reference:
K.Hiragi,
K.Nishio,
S.Moriyama,
T.Hamaguchi,
A.Mizoguchi,
K.Yonekura,
K.Tani,
T.Mizushima.
Structural Insights Into the Targeting Specificity of Ubiquitin Ligase For S. Cerevisiae Isocitrate Lyase But Not C. Albicans Isocitrate Lyase. J.Struct.Biol. V. 213 07748 2021.
ISSN: ESSN 1095-8657
PubMed: 34033899
DOI: 10.1016/J.JSB.2021.107748
Page generated: Wed Oct 2 20:34:09 2024
ISSN: ESSN 1095-8657
PubMed: 34033899
DOI: 10.1016/J.JSB.2021.107748
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1