Magnesium »
PDB 7k16-7kee »
7kc3 »
Magnesium in PDB 7kc3: X-Ray Structure of Lfa-1 I Domain Collected at 273 K
Protein crystallography data
The structure of X-Ray Structure of Lfa-1 I Domain Collected at 273 K, PDB code: 7kc3
was solved by
R.A.Woldeyes,
K.K.Hallenbeck,
S.J.Pfaff,
G.Lee,
S.V.Cortez,
M.J.Kelly,
K.Akassoglou,
M.R.Arkin,
J.S.Fraser,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.42 / 1.80 |
| Space group | P 32 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 104.896, 104.896, 51.644, 90, 90, 120 |
| R / Rfree (%) | 17.4 / 21.2 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the X-Ray Structure of Lfa-1 I Domain Collected at 273 K
(pdb code 7kc3). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the X-Ray Structure of Lfa-1 I Domain Collected at 273 K, PDB code: 7kc3:
In total only one binding site of Magnesium was determined in the X-Ray Structure of Lfa-1 I Domain Collected at 273 K, PDB code: 7kc3:
Magnesium binding site 1 out of 1 in 7kc3
Go back to
Magnesium binding site 1 out
of 1 in the X-Ray Structure of Lfa-1 I Domain Collected at 273 K
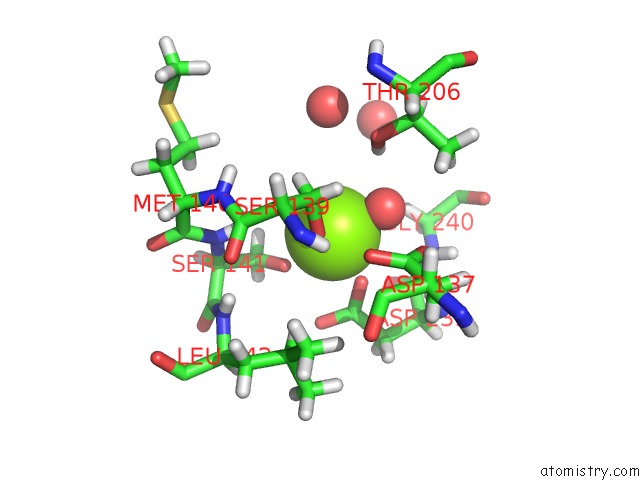
Mono view
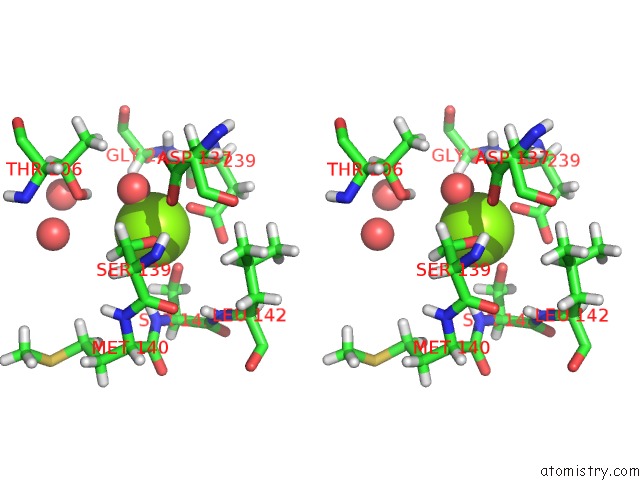
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of X-Ray Structure of Lfa-1 I Domain Collected at 273 K within 5.0Å range:
|
Reference:
R.A.Woldeyes,
K.K.Hallenbeck,
S.J.Pfaff,
G.Lee,
S.V.Cortez,
M.J.Kelly,
K.Akassoglou,
M.R.Arkin,
J.S.Fraser.
Divergent Conformational Dynamics Controls Allosteric Ligand Accessibility Across Evolutionarily Related I-Domain-Containing Integrins To Be Published.
Page generated: Wed Oct 2 22:10:59 2024
Last articles
Ca in 5V02Ca in 5UZW
Ca in 5UZU
Ca in 5UXA
Ca in 5UWM
Ca in 5UWK
Ca in 5UWL
Ca in 5UVE
Ca in 5UW5
Ca in 5UW6