Magnesium »
PDB 7ktn-7kz4 »
7ky7 »
Magnesium in PDB 7ky7: Structure of the S. Cerevisiae Phosphatidylcholine Flippase DNF2-LEM3 Complex in the Apo E1 State
Enzymatic activity of Structure of the S. Cerevisiae Phosphatidylcholine Flippase DNF2-LEM3 Complex in the Apo E1 State
All present enzymatic activity of Structure of the S. Cerevisiae Phosphatidylcholine Flippase DNF2-LEM3 Complex in the Apo E1 State:
7.6.2.1;
7.6.2.1;
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of the S. Cerevisiae Phosphatidylcholine Flippase DNF2-LEM3 Complex in the Apo E1 State
(pdb code 7ky7). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Structure of the S. Cerevisiae Phosphatidylcholine Flippase DNF2-LEM3 Complex in the Apo E1 State, PDB code: 7ky7:
In total only one binding site of Magnesium was determined in the Structure of the S. Cerevisiae Phosphatidylcholine Flippase DNF2-LEM3 Complex in the Apo E1 State, PDB code: 7ky7:
Magnesium binding site 1 out of 1 in 7ky7
Go back to
Magnesium binding site 1 out
of 1 in the Structure of the S. Cerevisiae Phosphatidylcholine Flippase DNF2-LEM3 Complex in the Apo E1 State
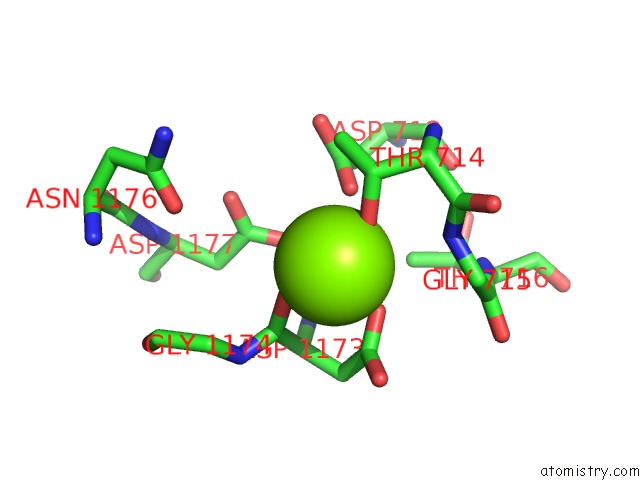
Mono view
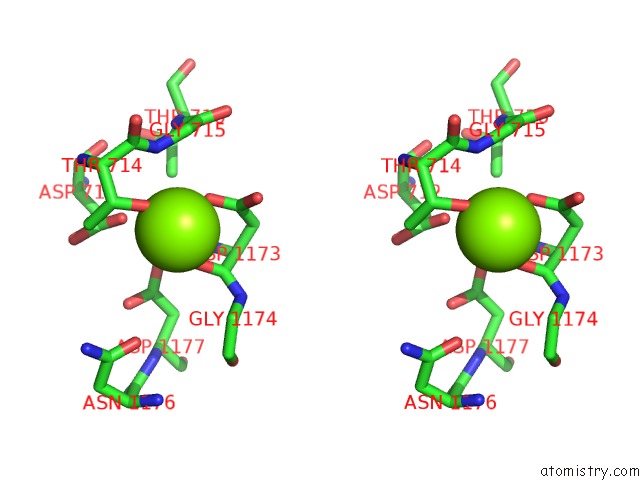
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of the S. Cerevisiae Phosphatidylcholine Flippase DNF2-LEM3 Complex in the Apo E1 State within 5.0Å range:
|
Reference:
L.Bai,
Q.You,
B.K.Jain,
H.D.Duan,
A.Kovach,
T.R.Graham,
H.Li.
Transport Mechanism of P4 Atpase Phosphatidylcholine Flippases. Elife V. 9 2020.
ISSN: ESSN 2050-084X
PubMed: 33320091
DOI: 10.7554/ELIFE.62163
Page generated: Thu Aug 14 08:46:21 2025
ISSN: ESSN 2050-084X
PubMed: 33320091
DOI: 10.7554/ELIFE.62163
Last articles
Mg in 7MY6Mg in 7MYB
Mg in 7MYC
Mg in 7MYA
Mg in 7MY9
Mg in 7MXW
Mg in 7MXB
Mg in 7MXK
Mg in 7MPP
Mg in 7MXJ