Magnesium »
PDB 7lcf-7lq2 »
7ljn »
Magnesium in PDB 7ljn: Structure of the Bradyrhizobium Diazoefficiens Cd-Ntase Cdng in Complex with Gtp
Protein crystallography data
The structure of Structure of the Bradyrhizobium Diazoefficiens Cd-Ntase Cdng in Complex with Gtp, PDB code: 7ljn
was solved by
A.Govande,
B.Lowey,
J.B.Eaglesham,
A.T.Whiteley,
P.J.Kranzusch,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.94 / 1.60 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 111.996, 68.687, 112.432, 90, 108.41, 90 |
| R / Rfree (%) | 19.7 / 21.8 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of the Bradyrhizobium Diazoefficiens Cd-Ntase Cdng in Complex with Gtp
(pdb code 7ljn). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Structure of the Bradyrhizobium Diazoefficiens Cd-Ntase Cdng in Complex with Gtp, PDB code: 7ljn:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Structure of the Bradyrhizobium Diazoefficiens Cd-Ntase Cdng in Complex with Gtp, PDB code: 7ljn:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 7ljn
Go back to
Magnesium binding site 1 out
of 4 in the Structure of the Bradyrhizobium Diazoefficiens Cd-Ntase Cdng in Complex with Gtp
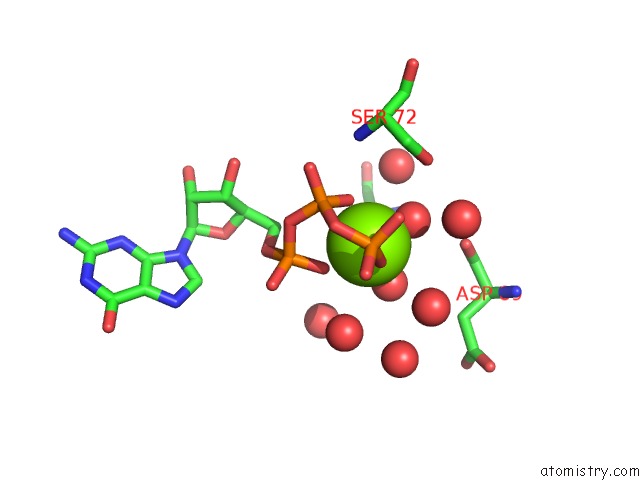
Mono view
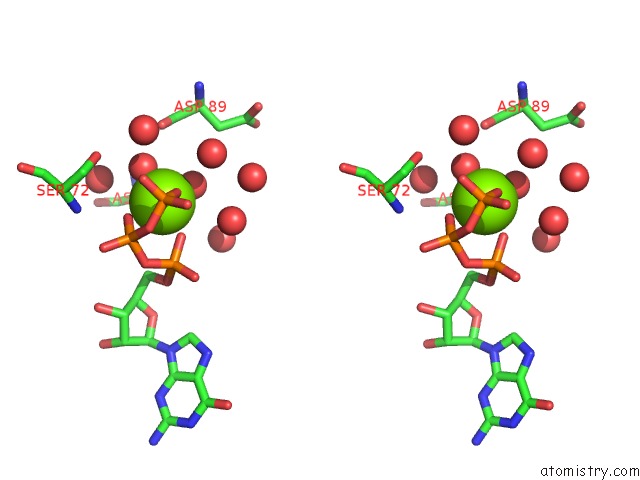
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of the Bradyrhizobium Diazoefficiens Cd-Ntase Cdng in Complex with Gtp within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 7ljn
Go back to
Magnesium binding site 2 out
of 4 in the Structure of the Bradyrhizobium Diazoefficiens Cd-Ntase Cdng in Complex with Gtp
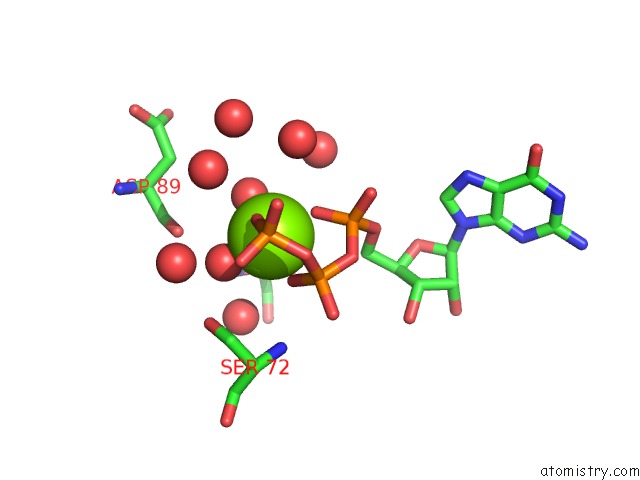
Mono view
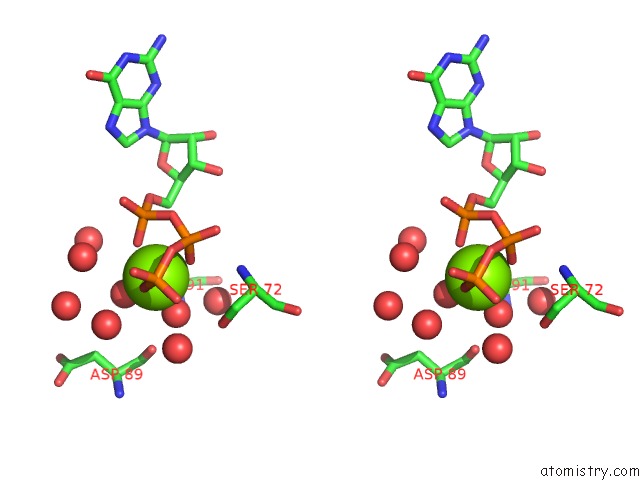
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of the Bradyrhizobium Diazoefficiens Cd-Ntase Cdng in Complex with Gtp within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 7ljn
Go back to
Magnesium binding site 3 out
of 4 in the Structure of the Bradyrhizobium Diazoefficiens Cd-Ntase Cdng in Complex with Gtp
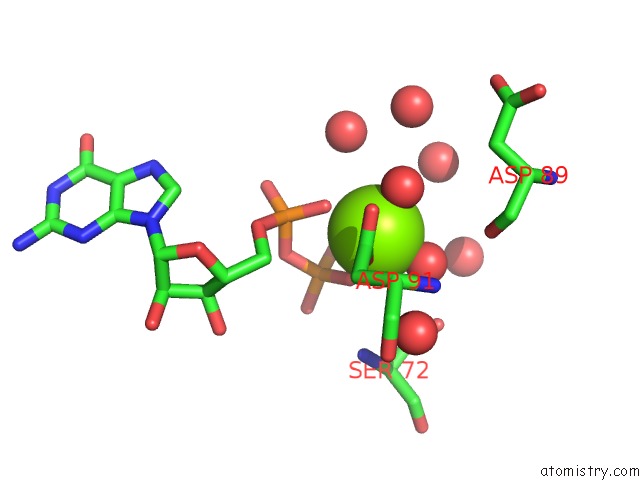
Mono view
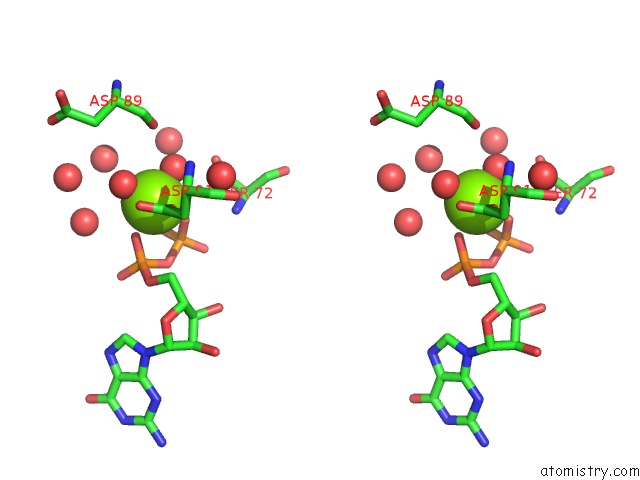
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structure of the Bradyrhizobium Diazoefficiens Cd-Ntase Cdng in Complex with Gtp within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 7ljn
Go back to
Magnesium binding site 4 out
of 4 in the Structure of the Bradyrhizobium Diazoefficiens Cd-Ntase Cdng in Complex with Gtp
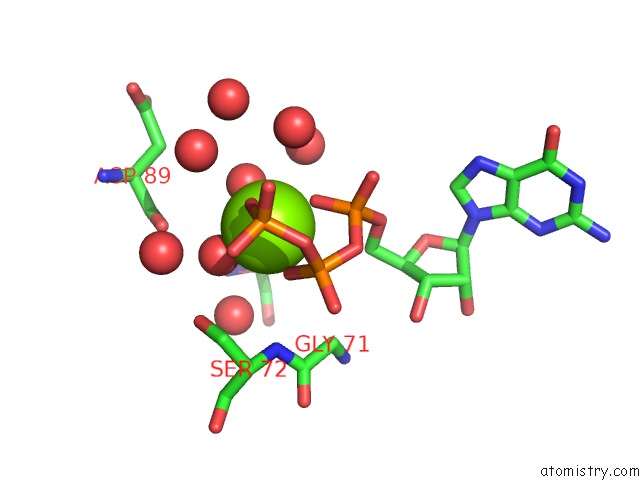
Mono view
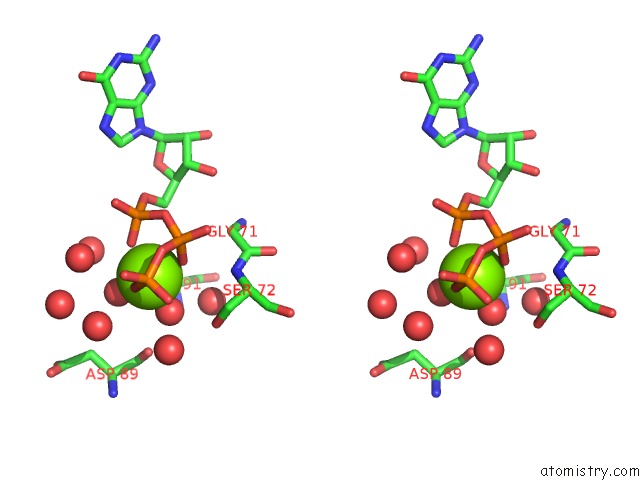
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Structure of the Bradyrhizobium Diazoefficiens Cd-Ntase Cdng in Complex with Gtp within 5.0Å range:
|
Reference:
A.A.Govande,
B.Duncan-Lowey,
J.B.Eaglesham,
A.T.Whiteley,
P.J.Kranzusch.
Molecular Basis of Cd-Ntase Nucleotide Selection in Cbass Anti-Phage Defense Cell Rep 2021.
ISSN: ESSN 2211-1247
Page generated: Wed Oct 2 23:25:36 2024
ISSN: ESSN 2211-1247
Last articles
Cl in 7UEYCl in 7UEV
Cl in 7UEX
Cl in 7UET
Cl in 7UER
Cl in 7UEM
Cl in 7UEL
Cl in 7UE0
Cl in 7UEQ
Cl in 7UEP