Magnesium »
PDB 7oup-7p8o »
7p84 »
Magnesium in PDB 7p84: Crystal Structure of L147A/I351A Variant of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii in Complex with Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine)
Enzymatic activity of Crystal Structure of L147A/I351A Variant of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii in Complex with Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine)
All present enzymatic activity of Crystal Structure of L147A/I351A Variant of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii in Complex with Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine):
2.5.1.6;
2.5.1.6;
Protein crystallography data
The structure of Crystal Structure of L147A/I351A Variant of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii in Complex with Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine), PDB code: 7p84
was solved by
E.Herrmann,
A.Peters,
N.V.Cornelissen,
A.Rentmeister,
D.Kuemmel,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.04 / 2.05 |
| Space group | P 32 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 76.104, 76.104, 280.674, 90, 90, 120 |
| R / Rfree (%) | 17.8 / 22.2 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of L147A/I351A Variant of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii in Complex with Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine)
(pdb code 7p84). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Crystal Structure of L147A/I351A Variant of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii in Complex with Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine), PDB code: 7p84:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Crystal Structure of L147A/I351A Variant of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii in Complex with Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine), PDB code: 7p84:
Jump to Magnesium binding site number: 1; 2; 3;
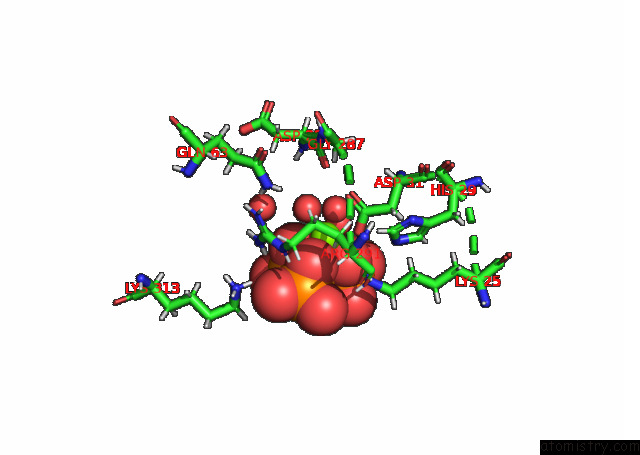
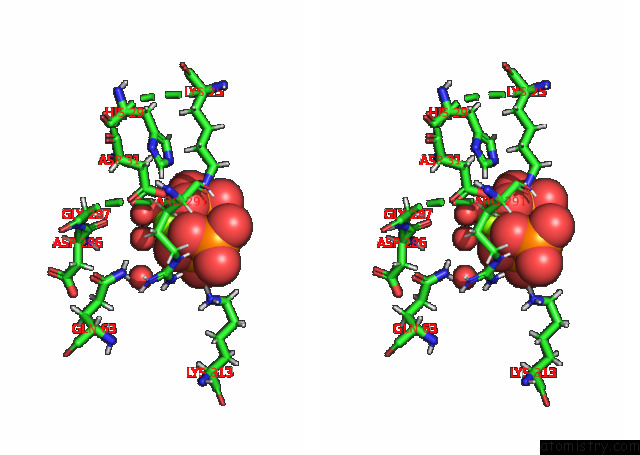
Magnesium binding site 1 out of 3 in 7p84
Go back to
Magnesium binding site 1 out
of 3 in the Crystal Structure of L147A/I351A Variant of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii in Complex with Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of L147A/I351A Variant of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii in Complex with Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine) within 5.0Å range:
|
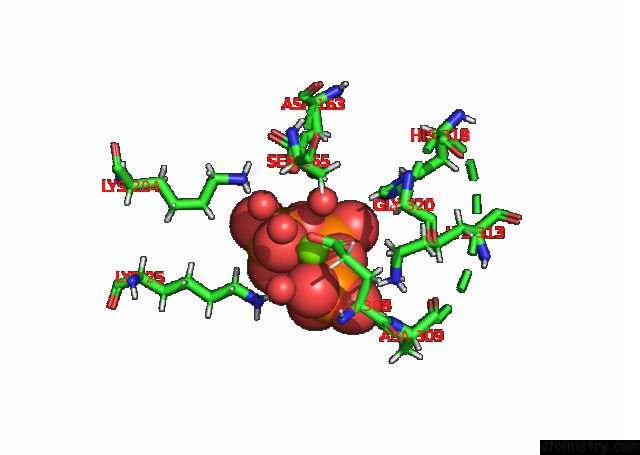
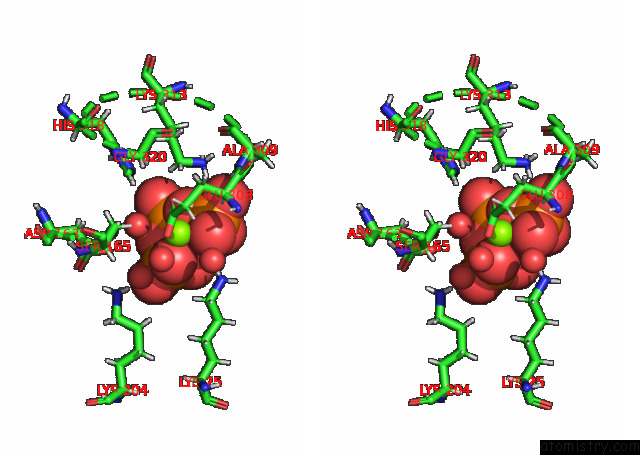
Magnesium binding site 2 out of 3 in 7p84
Go back to
Magnesium binding site 2 out
of 3 in the Crystal Structure of L147A/I351A Variant of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii in Complex with Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of L147A/I351A Variant of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii in Complex with Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine) within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 7p84
Go back to
Magnesium binding site 3 out
of 3 in the Crystal Structure of L147A/I351A Variant of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii in Complex with Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine)
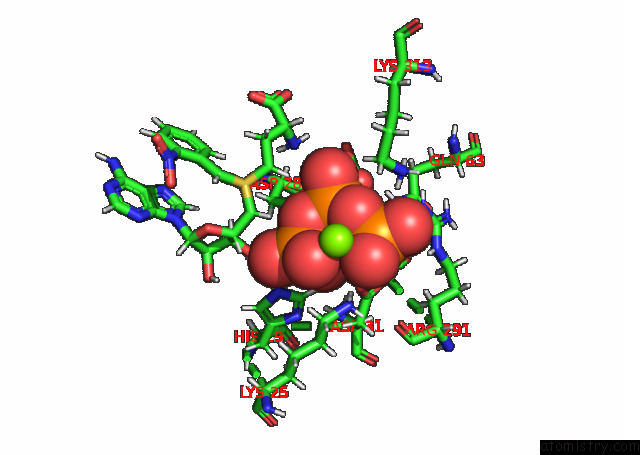
Mono view
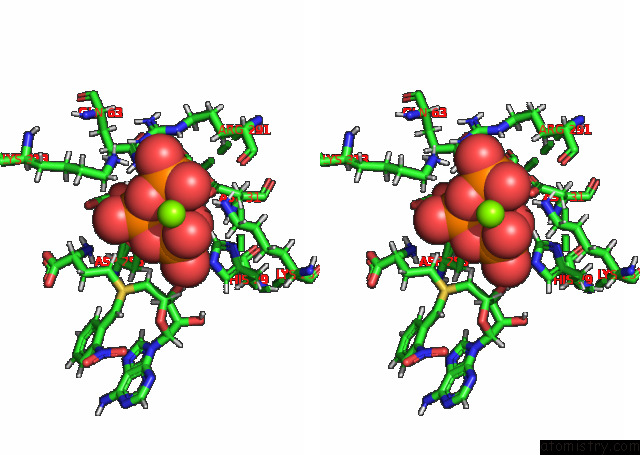
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of L147A/I351A Variant of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii in Complex with Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine) within 5.0Å range:
|
Reference:
A.Peters,
E.Herrmann,
N.V.Cornelissen,
N.Klocker,
D.Kummel,
A.Rentmeister.
Visible-Light Removable Photocaging Groups Accepted By Mjmat Variant: Structural Basis and Compatibility with Dna and Rna Methyltransferases. Chembiochem 2021.
ISSN: ESSN 1439-7633
PubMed: 34606675
DOI: 10.1002/CBIC.202100437
Page generated: Thu Aug 14 12:44:54 2025
ISSN: ESSN 1439-7633
PubMed: 34606675
DOI: 10.1002/CBIC.202100437
Last articles
Mg in 7S67Mg in 7S65
Mg in 7S66
Mg in 7S61
Mg in 7S60
Mg in 7S5Z
Mg in 7S5Y
Mg in 7S5X
Mg in 7S4X
Mg in 7RY4