Magnesium »
PDB 7p8v-7plh »
7pbn »
Magnesium in PDB 7pbn: Ruvab Branch Migration Motor Complexed to the Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset]
Enzymatic activity of Ruvab Branch Migration Motor Complexed to the Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset]
All present enzymatic activity of Ruvab Branch Migration Motor Complexed to the Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset]:
3.6.4.12;
3.6.4.12;
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Ruvab Branch Migration Motor Complexed to the Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset]
(pdb code 7pbn). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Ruvab Branch Migration Motor Complexed to the Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset], PDB code: 7pbn:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Ruvab Branch Migration Motor Complexed to the Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset], PDB code: 7pbn:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 7pbn
Go back to
Magnesium binding site 1 out
of 3 in the Ruvab Branch Migration Motor Complexed to the Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset]

Mono view
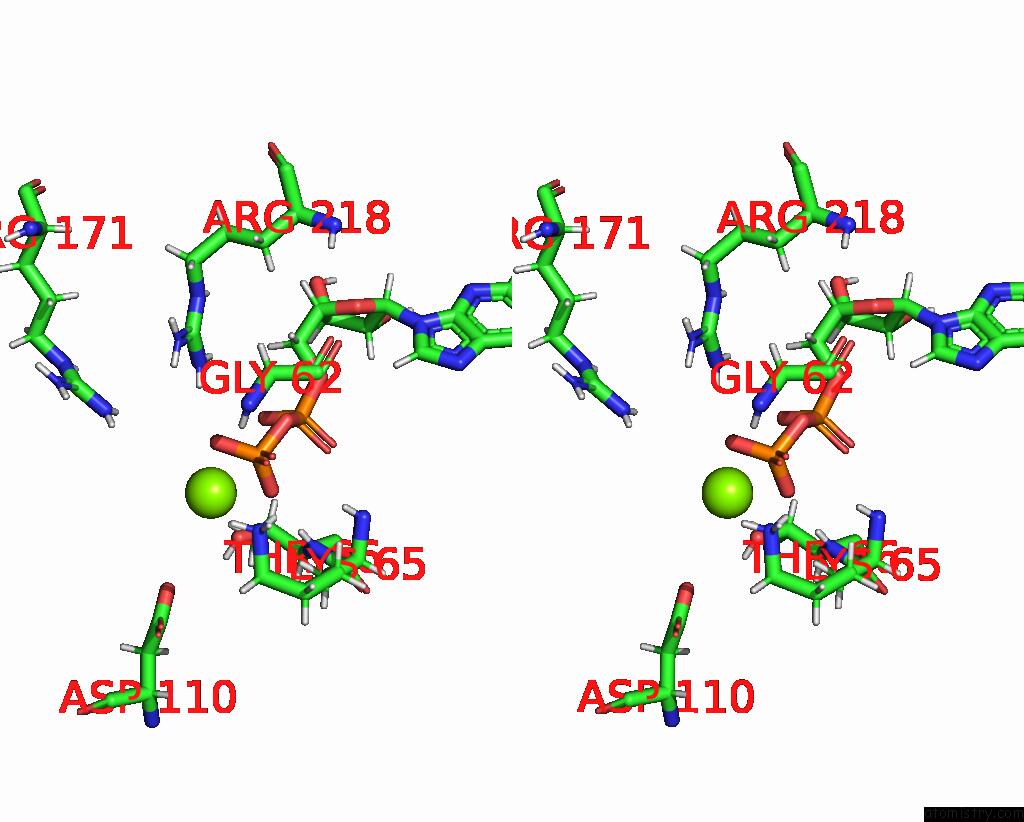
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Ruvab Branch Migration Motor Complexed to the Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset] within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 7pbn
Go back to
Magnesium binding site 2 out
of 3 in the Ruvab Branch Migration Motor Complexed to the Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset]

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Ruvab Branch Migration Motor Complexed to the Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset] within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 7pbn
Go back to
Magnesium binding site 3 out
of 3 in the Ruvab Branch Migration Motor Complexed to the Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset]
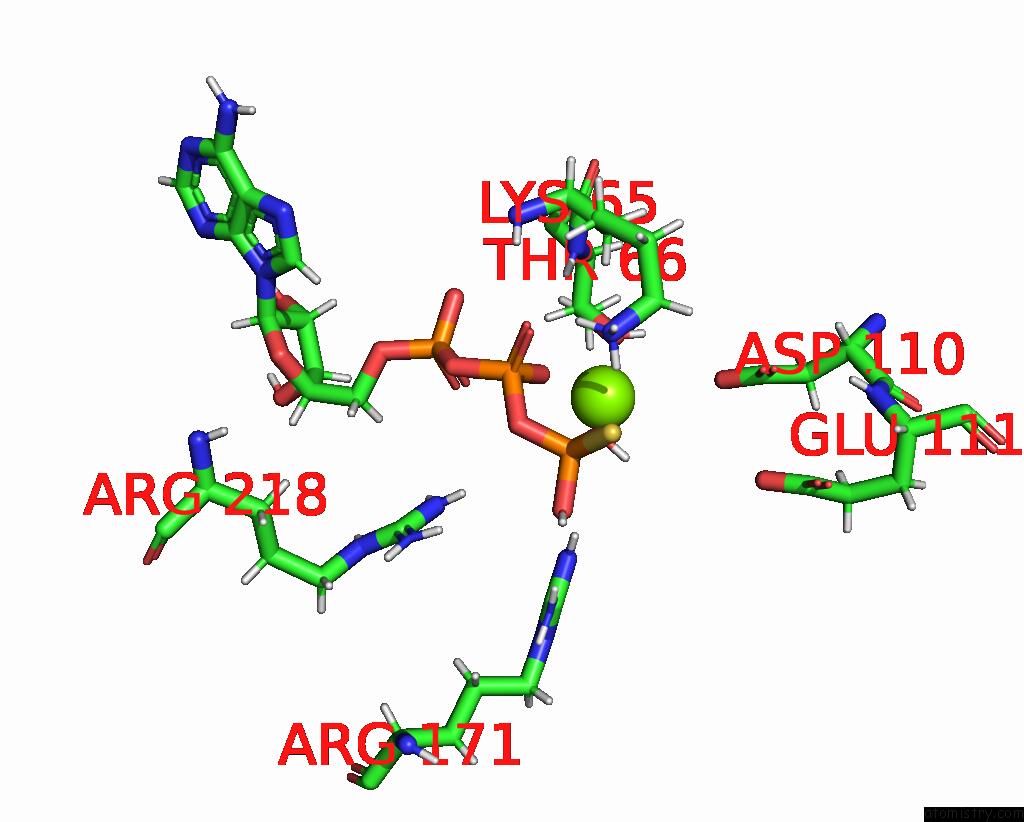
Mono view
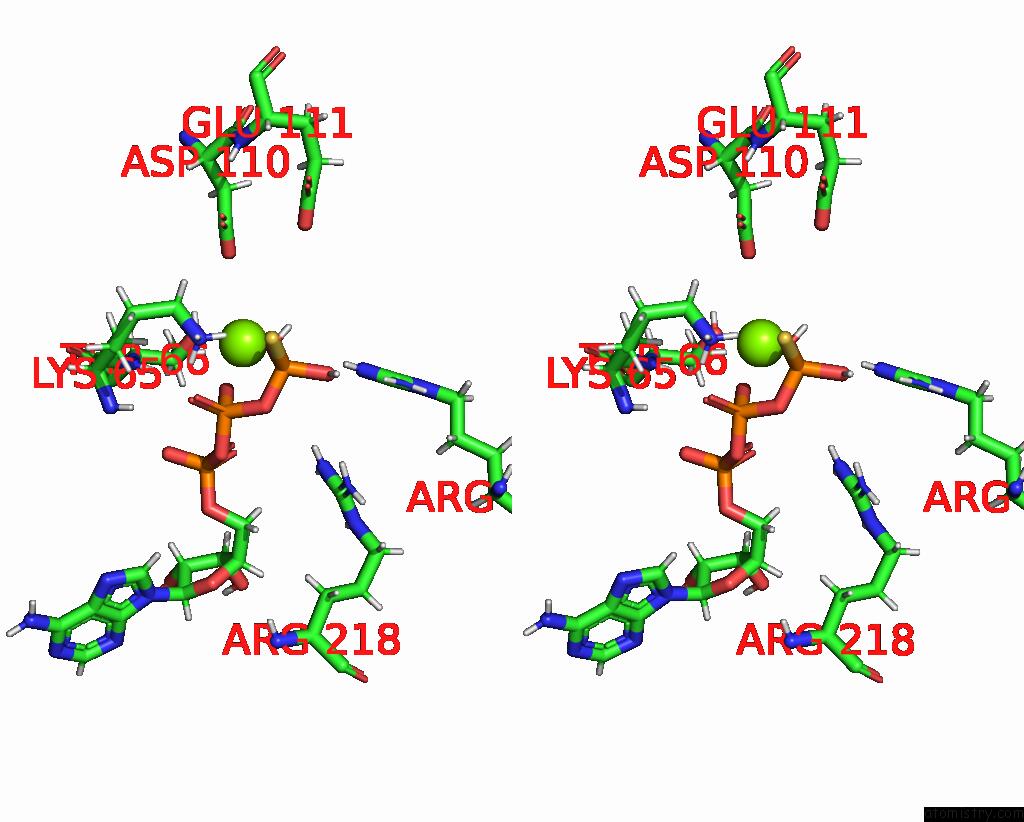
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Ruvab Branch Migration Motor Complexed to the Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset] within 5.0Å range:
|
Reference:
J.Wald,
D.Fahrenkamp,
N.Goessweiner-Mohr,
W.Lugmayr,
L.Ciccarelli,
O.Vesper,
T.C.Marlovits.
Mechanism of Aaa+ Atpase-Mediated Ruvab-Holliday Junction Branch Migration. Nature V. 609 630 2022.
ISSN: ESSN 1476-4687
PubMed: 36002576
DOI: 10.1038/S41586-022-05121-1
Page generated: Thu Aug 14 12:46:30 2025
ISSN: ESSN 1476-4687
PubMed: 36002576
DOI: 10.1038/S41586-022-05121-1
Last articles
Mg in 7S61Mg in 7S60
Mg in 7S5Z
Mg in 7S5Y
Mg in 7S5X
Mg in 7S4X
Mg in 7RY4
Mg in 7S5V
Mg in 7S2Y
Mg in 7S4F