Magnesium »
PDB 7p8v-7plh »
7pde »
Magnesium in PDB 7pde: Structure of Adenylyl Cyclase 9 in Complex with Gs Protein Alpha Subunit and Mant-Gtp
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of Adenylyl Cyclase 9 in Complex with Gs Protein Alpha Subunit and Mant-Gtp
(pdb code 7pde). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Structure of Adenylyl Cyclase 9 in Complex with Gs Protein Alpha Subunit and Mant-Gtp, PDB code: 7pde:
In total only one binding site of Magnesium was determined in the Structure of Adenylyl Cyclase 9 in Complex with Gs Protein Alpha Subunit and Mant-Gtp, PDB code: 7pde:
Magnesium binding site 1 out of 1 in 7pde
Go back to
Magnesium binding site 1 out
of 1 in the Structure of Adenylyl Cyclase 9 in Complex with Gs Protein Alpha Subunit and Mant-Gtp
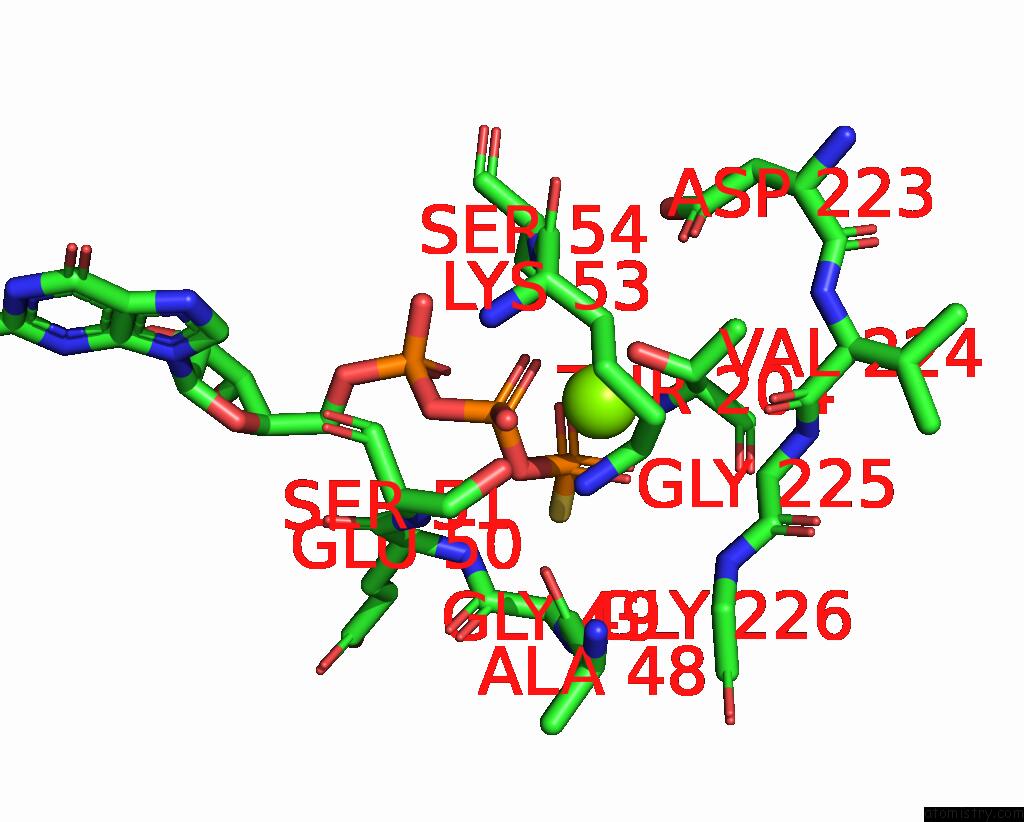
Mono view
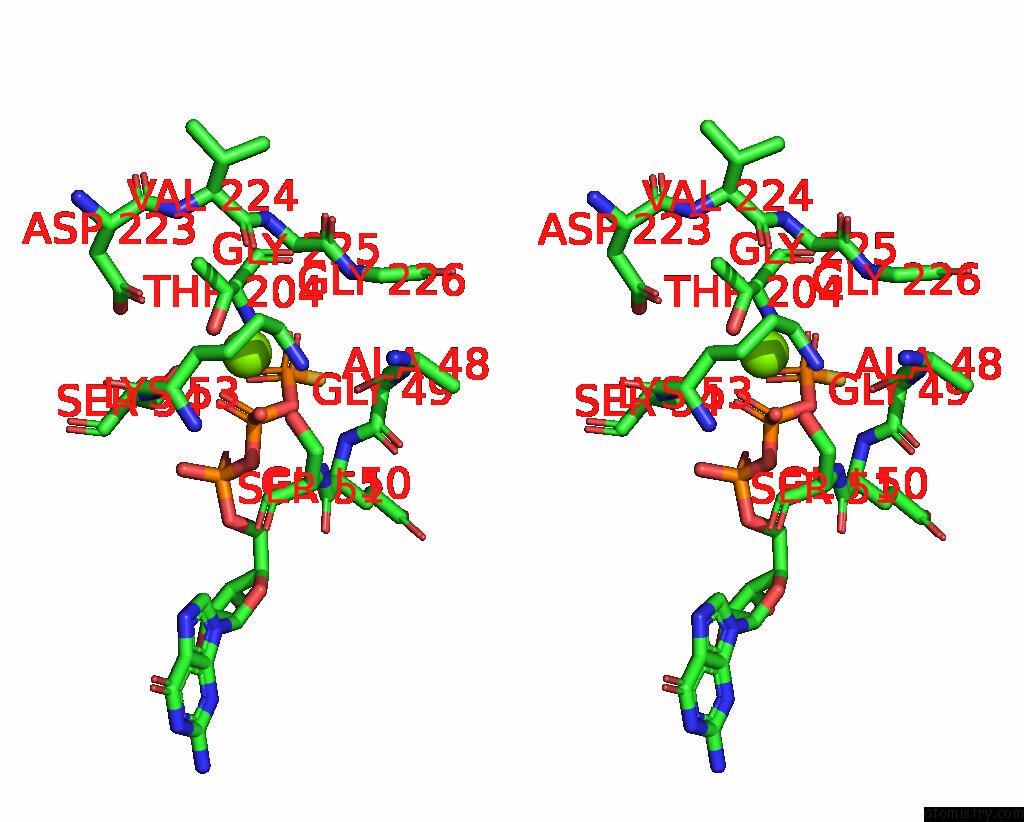
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of Adenylyl Cyclase 9 in Complex with Gs Protein Alpha Subunit and Mant-Gtp within 5.0Å range:
|
Reference:
C.Qi,
P.Lavriha,
V.Mehta,
B.Khanppnavar,
I.Mohammed,
Y.Li,
M.Lazaratos,
J.V.Schaefer,
B.Dreier,
A.Pluckthun,
A.N.Bondar,
C.W.Dessauer,
V.M.Korkhov.
Structural Basis of Adenylyl Cyclase 9 Activation. Nat Commun V. 13 1045 2022.
ISSN: ESSN 2041-1723
PubMed: 35210418
DOI: 10.1038/S41467-022-28685-Y
Page generated: Thu Aug 14 12:49:30 2025
ISSN: ESSN 2041-1723
PubMed: 35210418
DOI: 10.1038/S41467-022-28685-Y
Last articles
Mg in 7S67Mg in 7S65
Mg in 7S66
Mg in 7S61
Mg in 7S60
Mg in 7S5Z
Mg in 7S5Y
Mg in 7S5X
Mg in 7S4X
Mg in 7RY4