Magnesium »
PDB 7p8v-7plh »
7pds »
Magnesium in PDB 7pds: The Structure of PRIREP1 with Dsdna
Magnesium Binding Sites:
The binding sites of Magnesium atom in the The Structure of PRIREP1 with Dsdna
(pdb code 7pds). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the The Structure of PRIREP1 with Dsdna, PDB code: 7pds:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the The Structure of PRIREP1 with Dsdna, PDB code: 7pds:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 7pds
Go back to
Magnesium binding site 1 out
of 4 in the The Structure of PRIREP1 with Dsdna
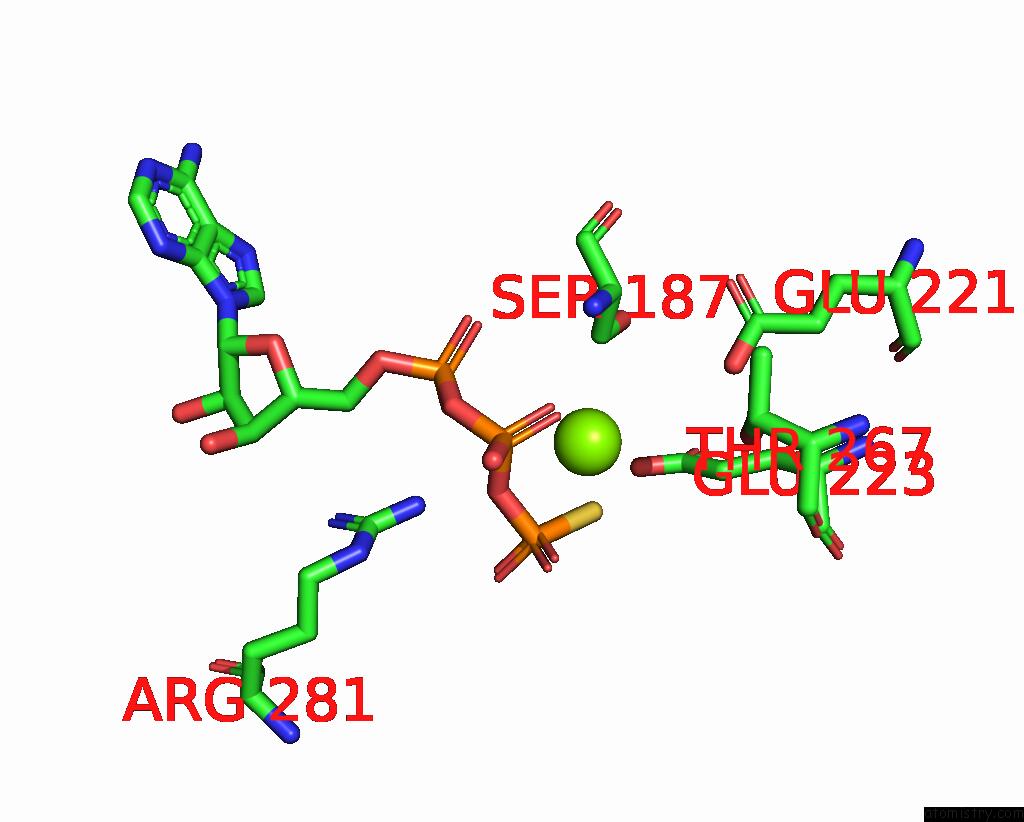
Mono view
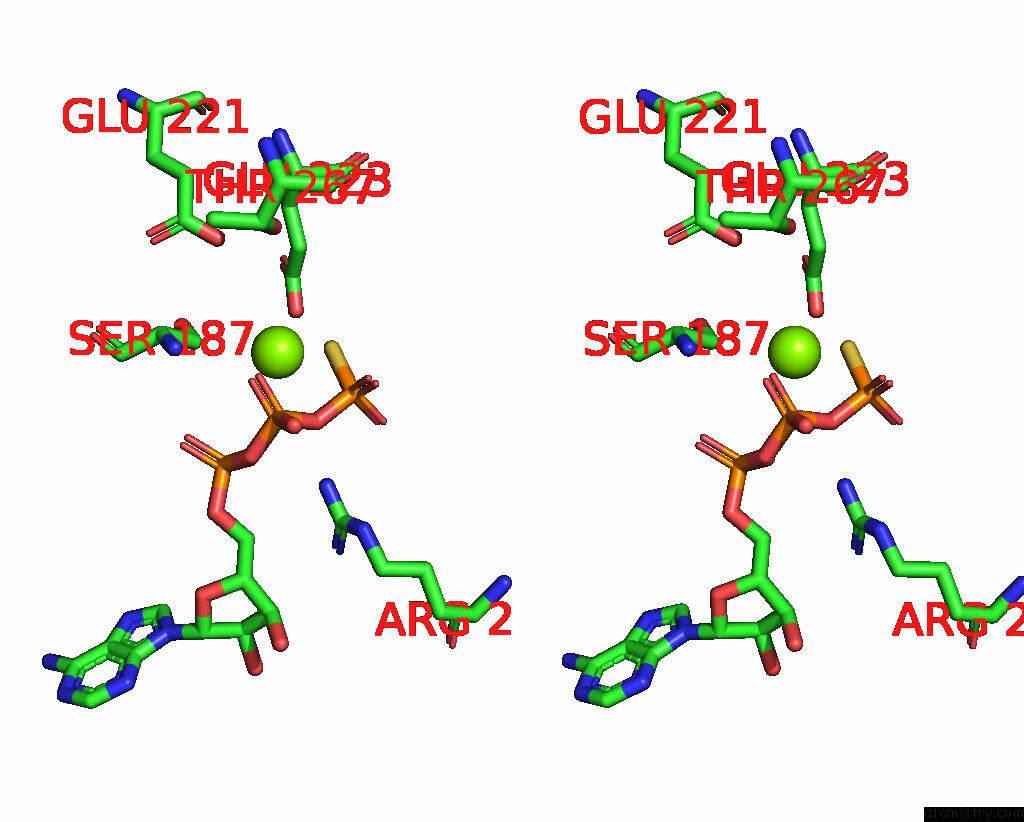
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of The Structure of PRIREP1 with Dsdna within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 7pds
Go back to
Magnesium binding site 2 out
of 4 in the The Structure of PRIREP1 with Dsdna
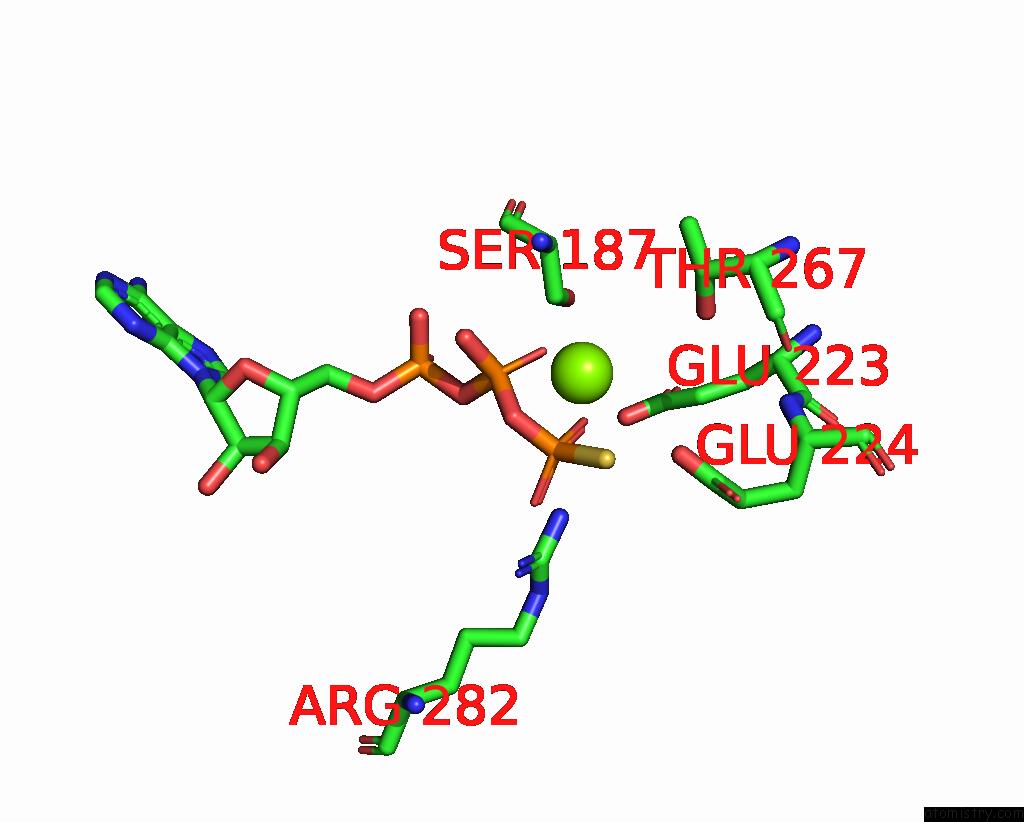
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of The Structure of PRIREP1 with Dsdna within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 7pds
Go back to
Magnesium binding site 3 out
of 4 in the The Structure of PRIREP1 with Dsdna
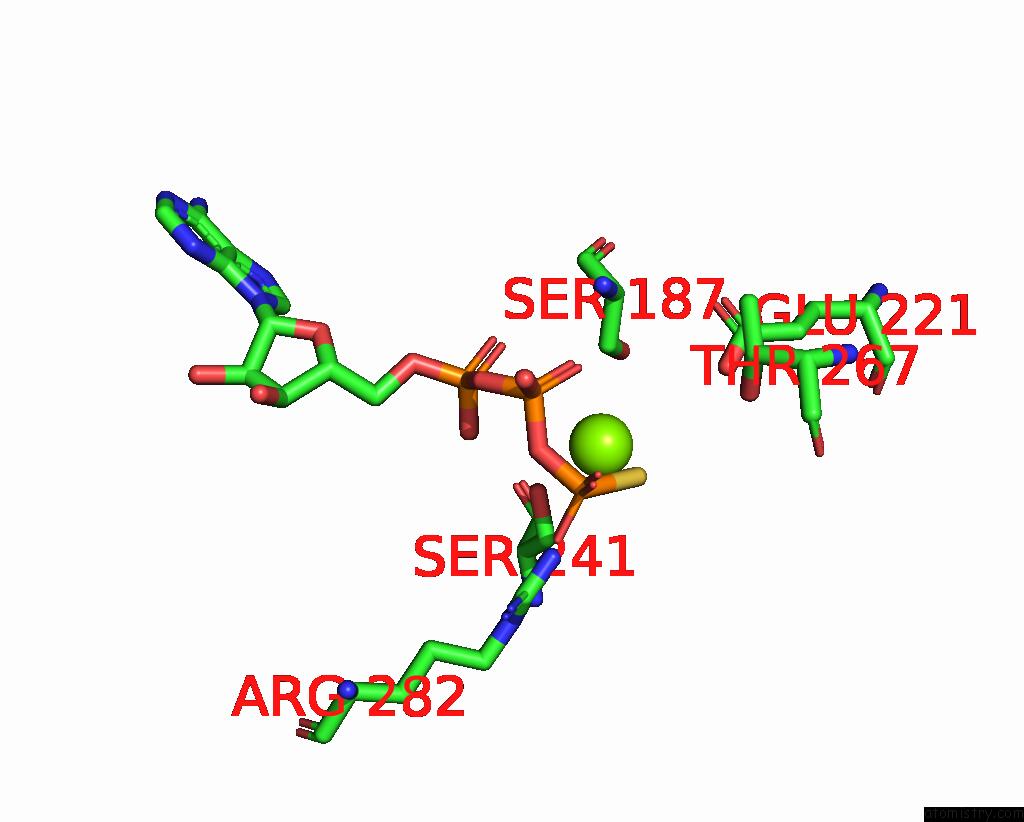
Mono view
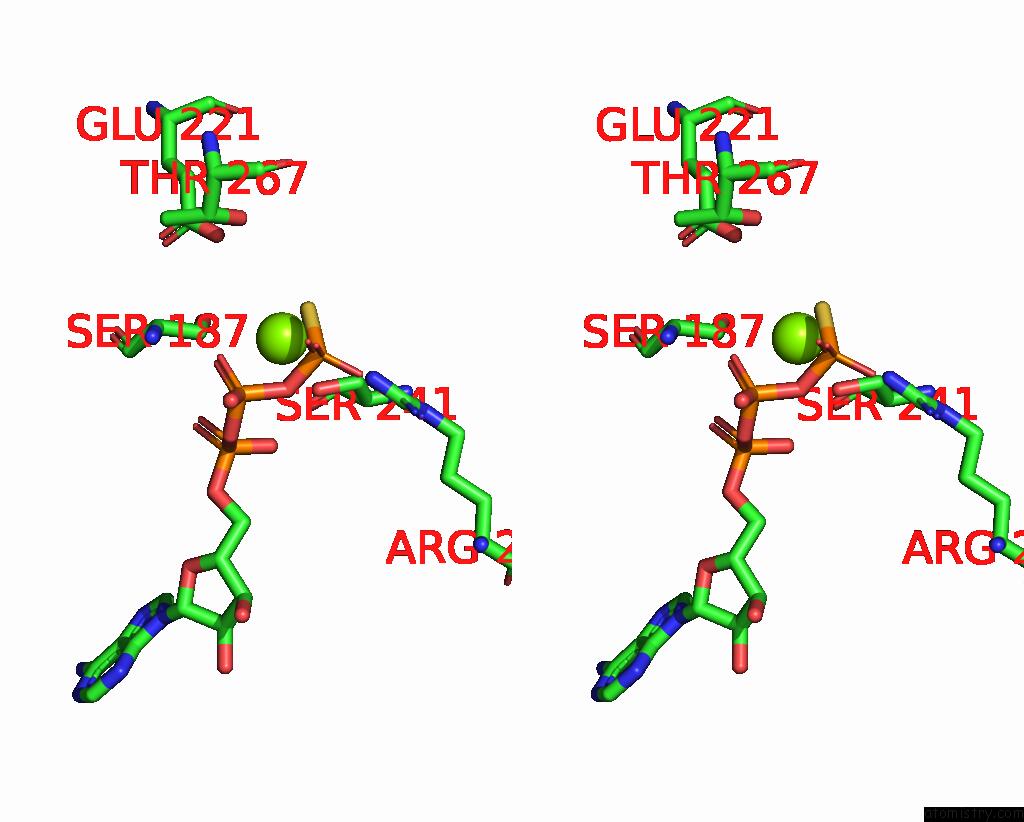
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of The Structure of PRIREP1 with Dsdna within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 7pds
Go back to
Magnesium binding site 4 out
of 4 in the The Structure of PRIREP1 with Dsdna
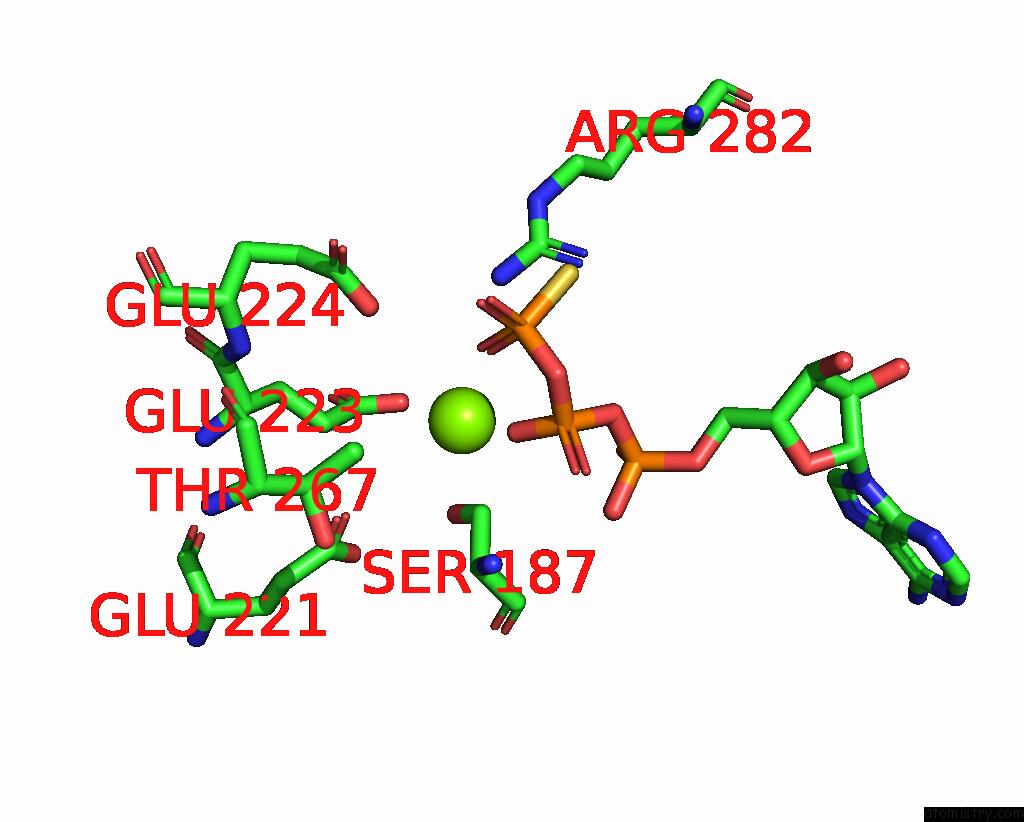
Mono view
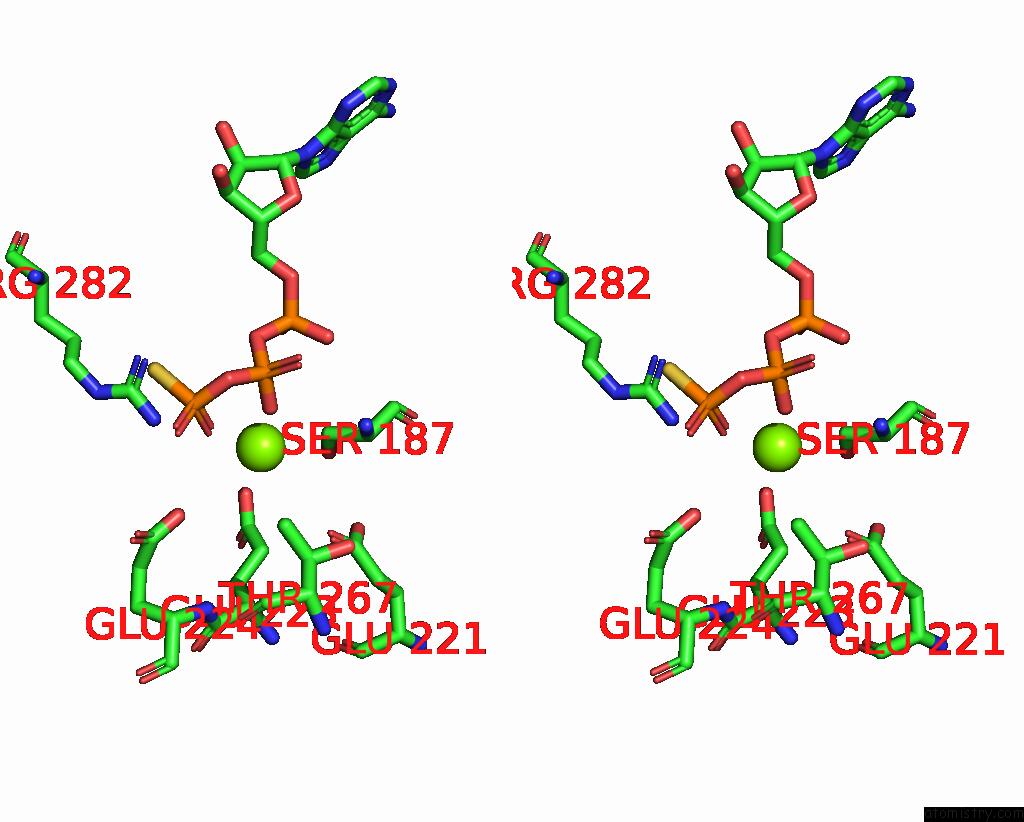
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of The Structure of PRIREP1 with Dsdna within 5.0Å range:
|
Reference:
C.Qiao,
G.Debiasi-Anders,
I.Mir-Sanchis.
Staphylococcal Self-Loading Helicases Couple the Staircase Mechanism with Inter Domain High Flexibility. Nucleic Acids Res. V. 50 8349 2022.
ISSN: ESSN 1362-4962
PubMed: 35871290
DOI: 10.1093/NAR/GKAC625
Page generated: Thu Aug 14 12:49:42 2025
ISSN: ESSN 1362-4962
PubMed: 35871290
DOI: 10.1093/NAR/GKAC625
Last articles
Mg in 7S67Mg in 7S65
Mg in 7S66
Mg in 7S61
Mg in 7S60
Mg in 7S5Z
Mg in 7S5Y
Mg in 7S5X
Mg in 7S4X
Mg in 7RY4